Team:MIT/Motivation
From 2012.igem.org
| Line 1: | Line 1: | ||
| - | Currently, the traditional method of synthetic biology uses proteins as on/off signals for cell based computing. The potential of these circuits is fundamentally limited by the number of proteins that can be orthogonally and simultaneously expressed. <p> | + | <p>Currently, the traditional method of synthetic biology uses proteins as on/off signals for cell based computing. The potential of these circuits is fundamentally limited by the number of proteins that can be orthogonally and simultaneously expressed. </p> |
| - | Imagine a circuit that senses for potential cancer cells and then produces a fluorescent protein that allows the cells to be easily identified by a doctor or surgeon. What would this require? <p> | + | <p>Imagine a circuit that senses for potential cancer cells and then produces a fluorescent protein that allows the cells to be easily identified by a doctor or surgeon. What would this require? </p> <p> |
| - | The first step, a cancer cell sensor, can be achieved by creating an mRNA sensor. The state of the art circuit with this function, (1 Multi-Input RNAi-Based Logic Circuit for Identification of Specific Cancer Cells. Xie et al. Science 2011) requires at least five composite parts for sensing high and low mRNA concentrations. The next part would need to process information from these five separate inputs, invert some of it, and send a signal to up regulate fluorescent protein production. This would require another set of promoters for each sensed mRNA and a repression system to produce the correct logic. The last step would be the induced expression of the signal protein; another unique promoter and protein pairing. [[File:Sensor.png]] | + | The first step, a cancer cell sensor, can be achieved by creating an mRNA sensor. The state of the art circuit with this function, (1 Multi-Input RNAi-Based Logic Circuit for Identification of Specific Cancer Cells. Xie et al. Science 2011) requires at least five composite parts for sensing high and low mRNA concentrations. The next part would need to process information from these five separate inputs, invert some of it, and send a signal to up regulate fluorescent protein production. This would require another set of promoters for each sensed mRNA and a repression system to produce the correct logic. The last step would be the induced expression of the signal protein; another unique promoter and protein pairing.</p> [[File:Sensor.png]] |
| - | < | + | <p> |
| - | + | This hypothetical circuit requires at least eleven unique promoters and proteins to function. At 1600 nucelotides per composite part, that means inserting 17,600 new bases into the cellular DNA. As shown in <whereever we got that graph from>, the current maximum number of promoters in a cellular circuit is six. [[File:Promotergraph.png]] </p> | |
| - | This hypothetical circuit requires at least eleven unique promoters and proteins to function. At 1600 nucelotides per composite part, that means inserting 17,600 new bases into the cellular DNA. As shown in <whereever we got that graph from>, the current maximum number of promoters in a cellular circuit is six. [[File:Promotergraph.png]] < | + | |
| - | + | ||
| + | <p> | ||
Clearly, a novel method of information processing is needed if we want to create complex circuits in vivo. For inspiration, we turned to "Scaling up digital circuit computation with DNA strand displacement cascades." Qian, L., Winfree, E. Science 2011. That research shows that it is possible to use the method of DNA strand displacement for complex circuits in vitro. The team built a circuit which calculated square roots with inputs of different short DNA strands representing binary numbers. The same hypothetical cancer sensing and highlighting circuit, designed using the strand displacement motif, requires only 3200 nucleotides of coding, creating over eighty strands which are roughly 40 bases in length. | Clearly, a novel method of information processing is needed if we want to create complex circuits in vivo. For inspiration, we turned to "Scaling up digital circuit computation with DNA strand displacement cascades." Qian, L., Winfree, E. Science 2011. That research shows that it is possible to use the method of DNA strand displacement for complex circuits in vitro. The team built a circuit which calculated square roots with inputs of different short DNA strands representing binary numbers. The same hypothetical cancer sensing and highlighting circuit, designed using the strand displacement motif, requires only 3200 nucleotides of coding, creating over eighty strands which are roughly 40 bases in length. | ||
[[File:Gategraph.png]] [[File:Calc.png]] | [[File:Gategraph.png]] [[File:Calc.png]] | ||
| - | < | + | </p> |
| - | + | ||
Integrating this into a ceullar system is non-trivial, but possible. | Integrating this into a ceullar system is non-trivial, but possible. | ||
Revision as of 15:00, 30 September 2012
Currently, the traditional method of synthetic biology uses proteins as on/off signals for cell based computing. The potential of these circuits is fundamentally limited by the number of proteins that can be orthogonally and simultaneously expressed.
Imagine a circuit that senses for potential cancer cells and then produces a fluorescent protein that allows the cells to be easily identified by a doctor or surgeon. What would this require?
The first step, a cancer cell sensor, can be achieved by creating an mRNA sensor. The state of the art circuit with this function, (1 Multi-Input RNAi-Based Logic Circuit for Identification of Specific Cancer Cells. Xie et al. Science 2011) requires at least five composite parts for sensing high and low mRNA concentrations. The next part would need to process information from these five separate inputs, invert some of it, and send a signal to up regulate fluorescent protein production. This would require another set of promoters for each sensed mRNA and a repression system to produce the correct logic. The last step would be the induced expression of the signal protein; another unique promoter and protein pairing.
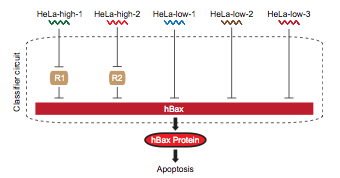
This hypothetical circuit requires at least eleven unique promoters and proteins to function. At 1600 nucelotides per composite part, that means inserting 17,600 new bases into the cellular DNA. As shown in <whereever we got that graph from>, the current maximum number of promoters in a cellular circuit is six. 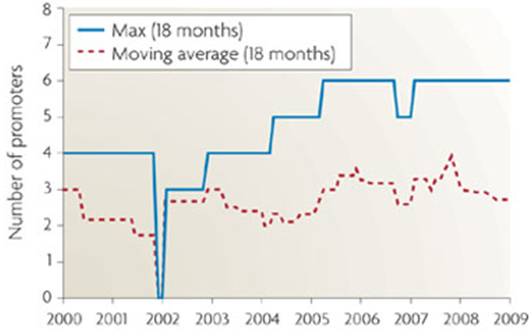
Clearly, a novel method of information processing is needed if we want to create complex circuits in vivo. For inspiration, we turned to "Scaling up digital circuit computation with DNA strand displacement cascades." Qian, L., Winfree, E. Science 2011. That research shows that it is possible to use the method of DNA strand displacement for complex circuits in vitro. The team built a circuit which calculated square roots with inputs of different short DNA strands representing binary numbers. The same hypothetical cancer sensing and highlighting circuit, designed using the strand displacement motif, requires only 3200 nucleotides of coding, creating over eighty strands which are roughly 40 bases in length.
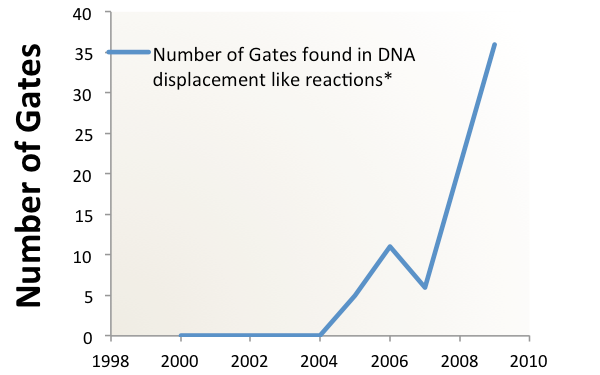
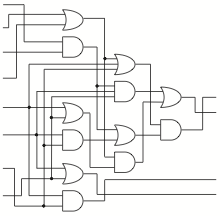
Integrating this into a ceullar system is non-trivial, but possible.
 "
"