Team:LMU-Munich/Data/gfp spore
From 2012.igem.org
Franzi.Duerr (Talk | contribs) |
|||
| (8 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
| - | <p align="justify">For creating fusion proteins for the '''Sporo'''beads, the genes ''gfp'', ''cotZ'' and ''cgeA'' were amplified and brought into Freiburg Standard. The restriction site NgoMIV was inserted just after the | + | <p align="justify">For creating fusion proteins for the '''Sporo'''beads, the genes ''gfp'', ''cotZ'' and ''cgeA'' were amplified and brought into Freiburg Standard. The restriction site NgoMIV was inserted just after the start codon of the genes of the spore crust proteins. Since this restriction site adds six additional base pairs, the resulting genes are two amino acids longer ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823032 CotZ], CgeA). Since there is no way of knowing if this insertion has any effect on protein expression, we created an additional truncated version. The deletion of the six base pairs downstream of the restriction site resulted in fusion proteins that were two amino acids shorter than the constructs above (derivatives of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823031 CotZ-2aa] and CgeA-2aa). |
| - | <br>Afterwards we first fused both versions of ''cotZ'' to its two native promoters, P<sub>''cotV''</sub> and P<sub>''cotYZ''</sub>, and to P<sub>''cgeA''</sub> | + | <br>Afterwards, we first fused both versions of ''cotZ'' to its two native promoters, P<sub>''cotV''</sub> and P<sub>''cotYZ''</sub>, and to P<sub>''cgeA''</sub>. For the two ''cgeA'' variants, we only used the native promoter P<sub>''cgeA''</sub>, and P<sub>''cotYZ''</sub>, the stronger one of the two promoters of the ''cotVWXYZ'' (for more details see [https://2012.igem.org/Team:LMU-Munich/Data/crustpromoters crust promotor evaluation]). In addition [http://partsregistry.org/Part:BBa_K823039 ''gfp''] was ligated to the terminator [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0014 B0014]. After sequence confirmation, we fused the ''cgeA''/''cotZ''- and ''gfp''-constructs together, applying the [http://partsregistry.org/Help:Assembly_standard_25 Freiburg standard] to create in-frame fusion proteins. This way, we created C-terminal ''gfp'' fusion to both spore crust proteins flanked by the promoters and terminator above. |
| - | <br> | + | <br>Since we want to display the fusion proteins on the surface of ''B. subtilis'' spores, we needed to clone our final constructs into an empty ''Bacillus'' vector, so that they would integrated into the chromosome of ''B. subtilis'' after transformation. We chose the empty vector pSB<sub>BS</sub>1C from our [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks '''''Bacillus''B'''io'''B'''rick'''B'''ox] for the ''cotZ'' constructs. |
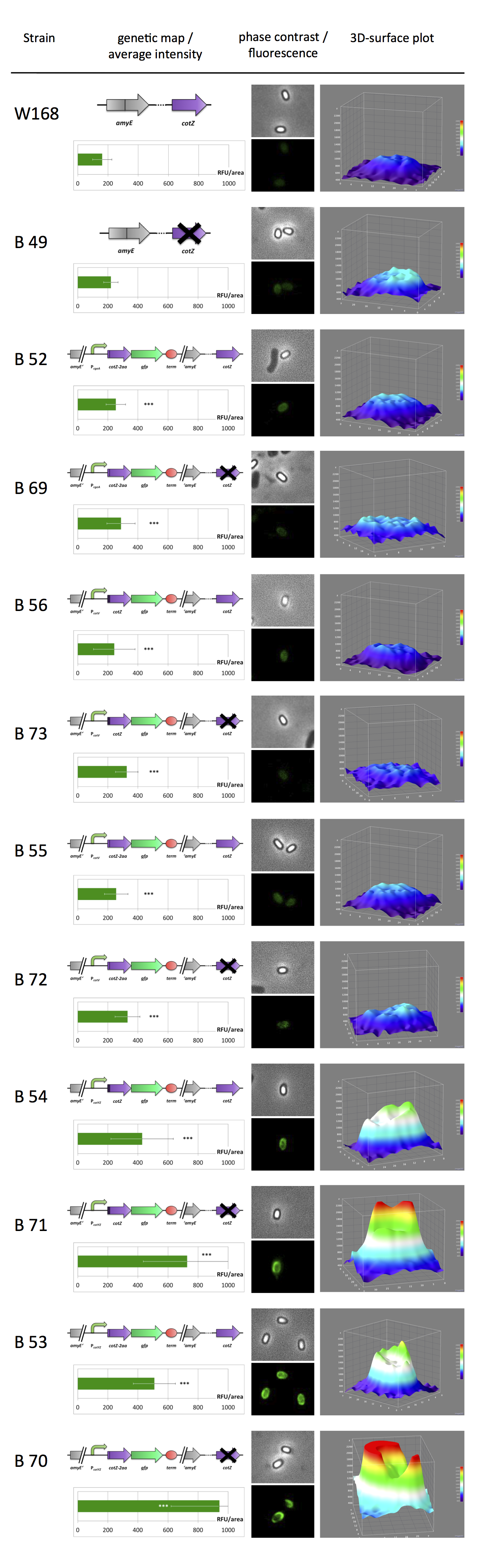
| - | <br>All | + | <br>All resulting '''Sporo'''beads were investigated by fluorescence microscopy. The intensity bar charts show the fluorescence intensity, while the 3D graphs illustrate the distribution of fluorescence intensity across the spore surface. This correlates with the localization of our fusion proteins in the crust. For image analysis we measured the fluorescence intensity of an area of 750 pixel per spore by using ImageJ and evaluated the results with the statistical software '''R'''. The graph below shows a summery of our results obtained from all constructs. |
</p> | </p> | ||
| - | |||
| - | <p align="justify">It appears the strain | + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" |
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[File:LMU gfp spore data.png|620px]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2">Fig. 1: Data of the various''' Sporo'''bead constructs</font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | |||
| + | <p align="justify">Fig. 1 verifies the previously described diffuse background fluorescence of wild type spores (W168). The B49 strain even shows an increased fluorescence background. Nevertheless there is a significant difference between gfp-'''Sporo'''beads and W168 and B 49, respectively. '''Sporo'''beads with present native ''cotZ'' revealed uniform distribution around the edge, whereas the ones with the deletion of ''cotZ'' elucidated a clear "polar" distribution. Additionally there is a difference in fluorescence activity between the two versions of ''cotZ'' recognizable. It appears the strain B 70 with the construct [http://partsregistry.org/Part:BBa_K823049 P<sub>''cotYZ''</sub>-''cotZ''<sub>-2aa</sub>-''gfp''-''terminator''] and the deleted native ''cotZ'' had the strongest fluorescence. But as it does not have a uniform fusion protein distribution, B 53 would be the strain for future '''Sporo'''bead applications. | ||
| + | <br>In some 3D graphs the picked cell does not represent the average fluorescence intensity depicted in the bar chart, because it was chosen randomly.</p> | ||
Latest revision as of 13:36, 26 October 2012
The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

GFP-Sporobead Evaluation
For creating fusion proteins for the Sporobeads, the genes gfp, cotZ and cgeA were amplified and brought into Freiburg Standard. The restriction site NgoMIV was inserted just after the start codon of the genes of the spore crust proteins. Since this restriction site adds six additional base pairs, the resulting genes are two amino acids longer ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823032 CotZ], CgeA). Since there is no way of knowing if this insertion has any effect on protein expression, we created an additional truncated version. The deletion of the six base pairs downstream of the restriction site resulted in fusion proteins that were two amino acids shorter than the constructs above (derivatives of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823031 CotZ-2aa] and CgeA-2aa).
Afterwards, we first fused both versions of cotZ to its two native promoters, PcotV and PcotYZ, and to PcgeA. For the two cgeA variants, we only used the native promoter PcgeA, and PcotYZ, the stronger one of the two promoters of the cotVWXYZ (for more details see crust promotor evaluation). In addition [http://partsregistry.org/Part:BBa_K823039 gfp] was ligated to the terminator [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0014 B0014]. After sequence confirmation, we fused the cgeA/cotZ- and gfp-constructs together, applying the [http://partsregistry.org/Help:Assembly_standard_25 Freiburg standard] to create in-frame fusion proteins. This way, we created C-terminal gfp fusion to both spore crust proteins flanked by the promoters and terminator above.
Since we want to display the fusion proteins on the surface of B. subtilis spores, we needed to clone our final constructs into an empty Bacillus vector, so that they would integrated into the chromosome of B. subtilis after transformation. We chose the empty vector pSBBS1C from our BacillusBioBrickBox for the cotZ constructs.
All resulting Sporobeads were investigated by fluorescence microscopy. The intensity bar charts show the fluorescence intensity, while the 3D graphs illustrate the distribution of fluorescence intensity across the spore surface. This correlates with the localization of our fusion proteins in the crust. For image analysis we measured the fluorescence intensity of an area of 750 pixel per spore by using ImageJ and evaluated the results with the statistical software R. The graph below shows a summery of our results obtained from all constructs.
|
Fig. 1 verifies the previously described diffuse background fluorescence of wild type spores (W168). The B49 strain even shows an increased fluorescence background. Nevertheless there is a significant difference between gfp-Sporobeads and W168 and B 49, respectively. Sporobeads with present native cotZ revealed uniform distribution around the edge, whereas the ones with the deletion of cotZ elucidated a clear "polar" distribution. Additionally there is a difference in fluorescence activity between the two versions of cotZ recognizable. It appears the strain B 70 with the construct [http://partsregistry.org/Part:BBa_K823049 PcotYZ-cotZ-2aa-gfp-terminator] and the deleted native cotZ had the strongest fluorescence. But as it does not have a uniform fusion protein distribution, B 53 would be the strain for future Sporobead applications.
In some 3D graphs the picked cell does not represent the average fluorescence intensity depicted in the bar chart, because it was chosen randomly.
 "
"