Team:Freiburg/Project/Overview
From 2012.igem.org
| Line 23: | Line 23: | ||
3. The third category of TALE assembly protocols applies Golden Gate Cloning (GGC)<sup>5,6,7,8,9</sup> (for details on GGC, see the [[Team:Freiburg/Project/Golden#GGC|Golden Gate Standard page]]). In all GGC-based TALE repeat assembly strategies, level 1 modules (i.e. single repeat gene fragments) are flanked by type IIs restriction sites adjacent to their first or last 4 nucleotides, respectively, that produce sticky ends after digestion with the type IIs restriction enzyme. Since each level 1 module codes for the same amino acid sequence (despite of the RVDs), the codon usage must be changed at these 4 external nucleotides for producing unique sticky ends that assemble in the predefined order after digestion. Consequently, the 4 bp overlaps of a level 1 module specify its future position within the TALE gene. | 3. The third category of TALE assembly protocols applies Golden Gate Cloning (GGC)<sup>5,6,7,8,9</sup> (for details on GGC, see the [[Team:Freiburg/Project/Golden#GGC|Golden Gate Standard page]]). In all GGC-based TALE repeat assembly strategies, level 1 modules (i.e. single repeat gene fragments) are flanked by type IIs restriction sites adjacent to their first or last 4 nucleotides, respectively, that produce sticky ends after digestion with the type IIs restriction enzyme. Since each level 1 module codes for the same amino acid sequence (despite of the RVDs), the codon usage must be changed at these 4 external nucleotides for producing unique sticky ends that assemble in the predefined order after digestion. Consequently, the 4 bp overlaps of a level 1 module specify its future position within the TALE gene. | ||
| - | So, in order to be able to target any sequence of DNA, a method that is using GGC requires N x | + | So, in order to be able to target any sequence of DNA, a method that is using GGC requires N x K modules. N signifies the number of level 1 module positions (i.e. number of modules that the TALE should contain after GGC) and K signifies the number of different repeats that the user should be able to put into each of the N positions (in most kits K equals 4, one repeat for each DNA base, see figure 1). |
Unfortunately, using GGC, only up to 10 modules <sup>5</sup> can be assembled with high accuracy. So in the GGC-based protocols, level 1 modules get assembled to form level 2 modules (oligorepeats). These level 2 modules need to be amplified and isolated before a second GGC reaction assembles them to form the complete repeat array. The bottleneck of the GGC-based methods is the need for amplification and isolation of level 2 modules, which costs a lot of time, requires some extra knowledge, additional enzymes and lab equipment (we actually tried one of the GGC-based open source kits, but, even after 2.5 weeks, were not able to assemble the whole TALE).<br><br> | Unfortunately, using GGC, only up to 10 modules <sup>5</sup> can be assembled with high accuracy. So in the GGC-based protocols, level 1 modules get assembled to form level 2 modules (oligorepeats). These level 2 modules need to be amplified and isolated before a second GGC reaction assembles them to form the complete repeat array. The bottleneck of the GGC-based methods is the need for amplification and isolation of level 2 modules, which costs a lot of time, requires some extra knowledge, additional enzymes and lab equipment (we actually tried one of the GGC-based open source kits, but, even after 2.5 weeks, were not able to assemble the whole TALE).<br><br> | ||
Revision as of 00:00, 27 October 2012
The GATE Assembly Kit
Therefore, we herein describe the Golden Gate cloning-based TAL Effector (GATE) Assembly platform, which enables literally everyone to produce low-cost, tailored TALEs within a few minutes of labwork and basic lab equipment. Moreover, we have automated this strategy and produced different TAL Effector Transcription Factors with 97 % success rate faster than any other method published before.
Review of existing TALE construction methods
Although TALE assembly is considerably easier than e.g. screening for novel zinc fingers, the highly repetitive structure of the TALE gene implies some challenges, because conventional PCR or homologous recombination-based gene assembly strategies cannot be applied. To our knowledge, the numerous approaches of TAL-Effector gene assembly, published so far, fall under the following three categories:
1. Few groups have applied methods called unit assembly1 or Restriction Enzyme And Ligation (REAL)2. In the first step, both strategies perform conventional restriction enzyme digestion in order to assemble two gene fragments of single repeats. The pairs of repeat gene fragments are subsequently assembled to form tetramers, and this highly hierarchical assembly strategy is continued until the desired number of repeats is assembled. These platforms obviously involve multiple laborious and time consuming rounds of digestion, ligation and isolation of the right ligation products. The recently published fast ligation-based automatable solid-phase high-throughput (FLASH) system circumvents major challenges of REAL by attaching the first repeat to streptavidin-coated magnetic beads and, successively, adding further repeats or oligorepeats from a 376-plasmid library. Although Reyon et al.11 claim that FLASH can also be performed manually, this probably does not represent the most convenient and low-cost protocol for iGEM students.
2. We call the second category of TALE production methods the synthesis optimization approach. The major challenge of TAL synthesis is the highly repetitive amino acid sequence of the DNA binding part. Since synthetic genes are typically produced from overlapping synthesized oligos, overlaps of different pairs of overlapping oligos need to be distinct. The synthesis optimization approach employs a sophisticated computer program that optimizes codon usage in order to reduce repetitiveness of the TAL gene and calculates optimal oligos for synthesis3,4. Although this approach might be the method of the future, it is currently too expensive for iGEM teams.
3. The third category of TALE assembly protocols applies Golden Gate Cloning (GGC)5,6,7,8,9 (for details on GGC, see the Golden Gate Standard page). In all GGC-based TALE repeat assembly strategies, level 1 modules (i.e. single repeat gene fragments) are flanked by type IIs restriction sites adjacent to their first or last 4 nucleotides, respectively, that produce sticky ends after digestion with the type IIs restriction enzyme. Since each level 1 module codes for the same amino acid sequence (despite of the RVDs), the codon usage must be changed at these 4 external nucleotides for producing unique sticky ends that assemble in the predefined order after digestion. Consequently, the 4 bp overlaps of a level 1 module specify its future position within the TALE gene.
So, in order to be able to target any sequence of DNA, a method that is using GGC requires N x K modules. N signifies the number of level 1 module positions (i.e. number of modules that the TALE should contain after GGC) and K signifies the number of different repeats that the user should be able to put into each of the N positions (in most kits K equals 4, one repeat for each DNA base, see figure 1).
Unfortunately, using GGC, only up to 10 modules 5 can be assembled with high accuracy. So in the GGC-based protocols, level 1 modules get assembled to form level 2 modules (oligorepeats). These level 2 modules need to be amplified and isolated before a second GGC reaction assembles them to form the complete repeat array. The bottleneck of the GGC-based methods is the need for amplification and isolation of level 2 modules, which costs a lot of time, requires some extra knowledge, additional enzymes and lab equipment (we actually tried one of the GGC-based open source kits, but, even after 2.5 weeks, were not able to assemble the whole TALE).
GATE Assembly Kit
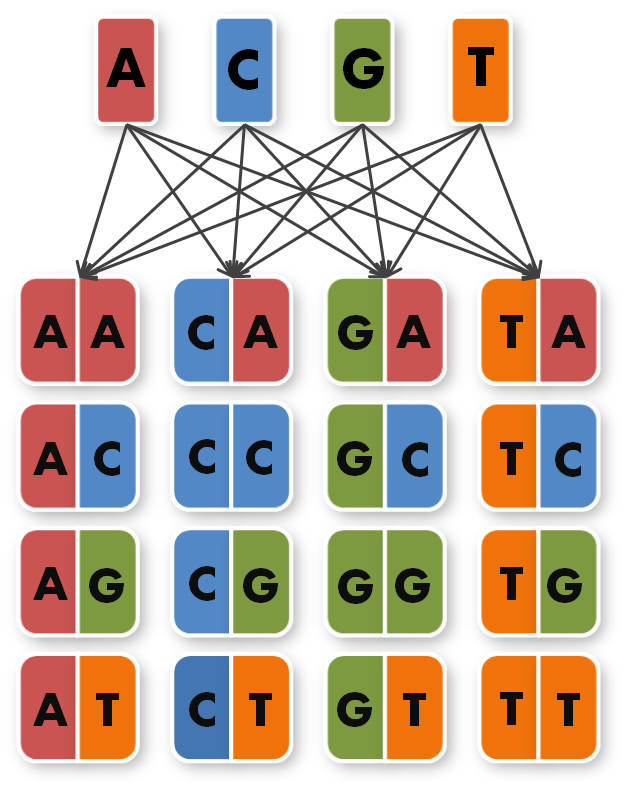
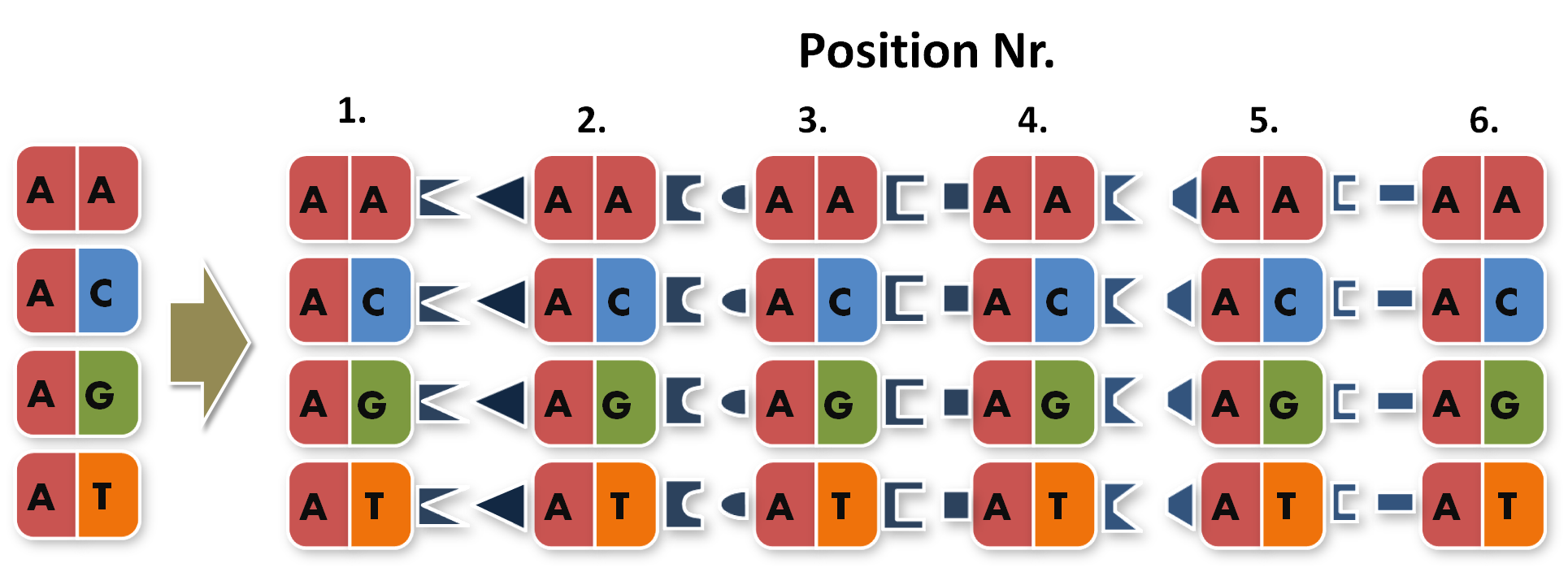
So we needed to further reduce N down to 6 to obtain a reasonable toolkit size of 96 level 1 modules. We actually liked the idea that our kit would perfectly fit on a 96 well plate.
Next, we looked into the literature to check, if TALEs that recognize 14 bp (instead of around 18 bp) are actually functional. We were very fortunate to see that efficiency of TAL transcription factors (TAL-TFs) 10 and TAL effector nucleases (TALENs)11 remains constant between for target sequences between 13 and 20 bp. Moreover, Zhang et al. published splendid results with 14 bp-binding TAL-TFs in a human cell line7.
Since we wanted our TALEs to function in both bacteria and eukaryotic systems, while published TAL repeats were always designed for one particular organism, we decided to design the direpeat nucleotide sequences from scratch: We used the amino acid sequence of the hex3 gene of Xanthomonas oryzae to find out the amino acid sequences for the 16 direpeats. Next, we reverse-tanslated the sequences into DNA, codon optimized them for E.coli and human cells and reduced homologies between and within gene fragments (only the extention PCR binding sites were the same for every direpeat gene).
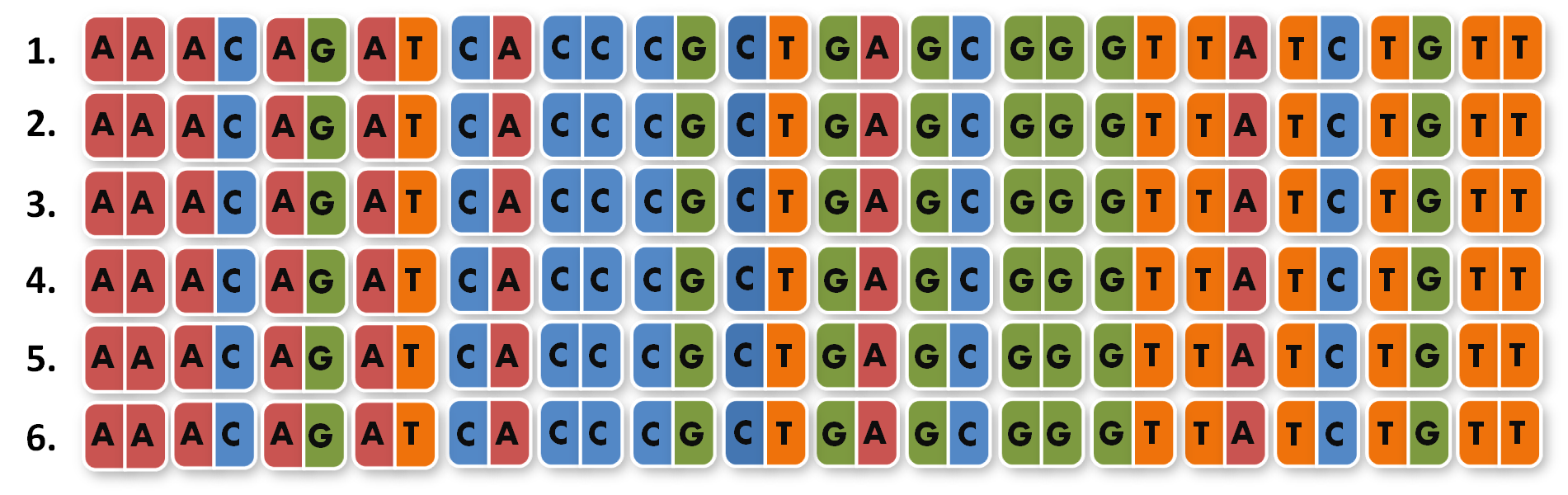
After receiving the sequences that were synthesized as G-blocks by IDT, we performed 6 extention PCRs on every sequence to add 4 bp overlaps, BsmBI restriction sites and iGEM prefix and suffix to the parts. The 4 bp overlaps would later determine the position of the respective direpeat in the repeat array of the TALE.
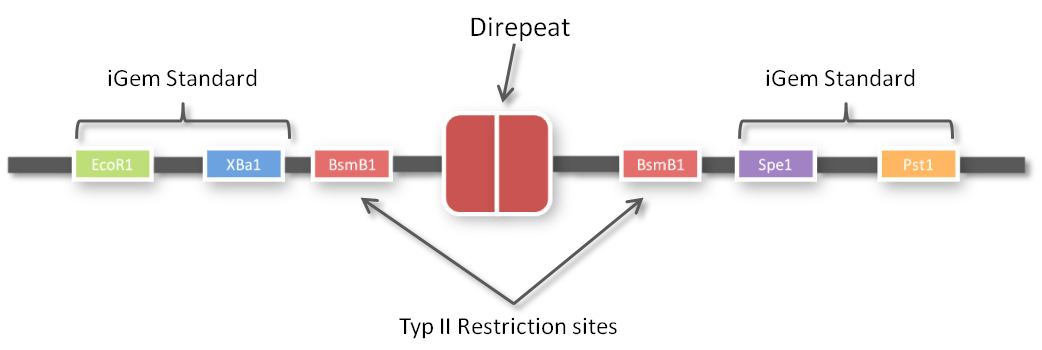
One of the advantages of GGC is that you can insert complete plasmids containing the parts you want to assemble. So we decided to clone all 96 parts into the standard iGEM vector pSB1C3. We hypothesized that the BsmBI restriction site in the chloramphenicol gene would decrease GGC efficiency, so we performed a mutagenesis PCR to introduce the silent mutation (G434C) prior to cloning the 96 PCR products into it. When doing so many cloning experiments at a time, error rate needs to be minimal, so at first, we spent weeks optimizing every single step from the G-block to the Golden Gate standard compatible BioBrick (see protocol section). In the end, we are very happy that we have a full GATE assembly kit with 96 unique direpeats and 100% accurate sequencing results.
Our first attempts to use the GATE assembly kit were actually very discouraging - no colonies were found on the agar plates after transforming the GGC product into DH10B cells for more than one week - at least, we knew that our ccdb kill cassette was working well (details about the expression vector). After systematically testing all kinds of buffers and reaction additives, the results where quite overwhelming. We were even able to dramatically reduce GGC reaction time down to 2.5 hours - which is probably the fastest way anyone has ever built a custom tal effector.
References
1. Huang, P. et al. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 29, 699–700 (2011).
2. Sander, J. D. et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol 29, 697–698 (2011).
3. Hoover, D. M. & Lubkowski, J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucl Acids Res 30, e43–e43 (2002).
4. Miller, J. C. et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29, 143–148 (2010).
5. Morbitzer, R., Elsaesser, J., Hausner, J. & Lahaye, T. Assembly of Custom TALE-Type DNA Binding Domains by Modular Cloning. Nucl Acids Res 39, 5790–5799 (2011).
6. Weber, E., Gruetzner, R., Werner, S., Engler, C. & Marillonnet, S. Assembly of designer TAL effectors by golden gate cloning. PloS one 6, e19722 (2011).
7. Zhang, F. et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29, 149–153 (2011).
8. Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucl Acids Res 39, e82 (2011).
9. Li, T. et al. Modularly Assembled Designer TAL Effector Nucleases for Targeted Gene Knockout and Gene Replacement in Eukaryotes. Nucl Acids Res 39, 6315–6325 (2011).
10. Boch, J. et al. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 326, 1509–1512 (2009).
11. Reyon, D. et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30, 460–465 (2012).
 "
"
