Team:Cambridge/Biologger/Results
From 2012.igem.org
(→Biologger Results) |
|||
| Line 70: | Line 70: | ||
Having our instrumentation completed, as can be seen in our [[Team:Cambridge/Biologger/Overview|<span style="color:#000066"><u>Instrumentation (Biologger) page,</u></span>]] the sensitivity of the sensor placed in the right position was tested using a dilution series of luciferase-producing E.coli. 20ml Cultures were grown overnight from single colonies. The cultures were induced with 40ul of 1.5M arabinose (for a final concentration of 3mM). | Having our instrumentation completed, as can be seen in our [[Team:Cambridge/Biologger/Overview|<span style="color:#000066"><u>Instrumentation (Biologger) page,</u></span>]] the sensitivity of the sensor placed in the right position was tested using a dilution series of luciferase-producing E.coli. 20ml Cultures were grown overnight from single colonies. The cultures were induced with 40ul of 1.5M arabinose (for a final concentration of 3mM). | ||
Cultures were left for 2 1/2 hours for full induction. Subsequently, a culture was pelleted and resuspended in 4ml LB. Doubling dilutions, of volume 2ml, were made from this concentrate, down to 1/8th concentration. | Cultures were left for 2 1/2 hours for full induction. Subsequently, a culture was pelleted and resuspended in 4ml LB. Doubling dilutions, of volume 2ml, were made from this concentrate, down to 1/8th concentration. | ||
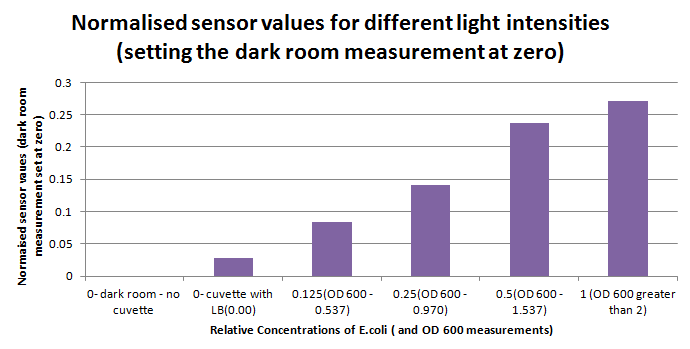
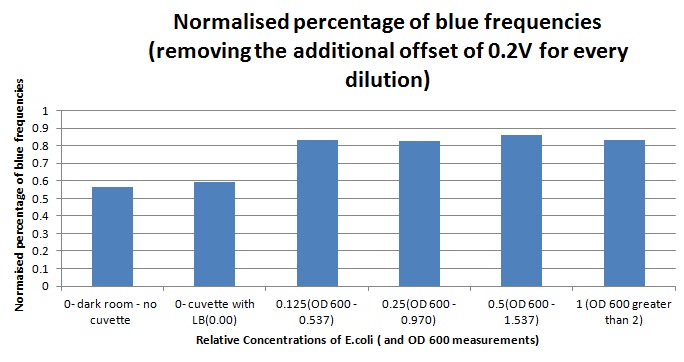
| - | 1ml of each 2ml dilution was analysed in each cuvette, which was placed in the cuvette holder we made ourselves. The result was very good. An almost linear relationship was obtained when data were normalised with the sensor value taken in the dark room (the latter set at zero) without using the cuvette holder (1-(sensor value/sensor value in absolute dark)), presenting the sensitivity of the sensor to different intensities of light. This behaviour was expected due to the changing offset affecting the luciferase spectrum curve at different light intensities. The offset, using our data, was calculated to be about 0.2V for each dilution. A second graph is shown which takes into account this offset (and removes it), thus showing the presence of blue frequencies. The result was as expected, as the presence of blue frequencies throughout the dilution series is not only detected, but also found to be approximately constant. The raw data of this investigation can be found in the [[Team:Cambridge/ | + | 1ml of each 2ml dilution was analysed in each cuvette, which was placed in the cuvette holder we made ourselves. The result was very good. An almost linear relationship was obtained when data were normalised with the sensor value taken in the dark room (the latter set at zero) without using the cuvette holder (1-(sensor value/sensor value in absolute dark)), presenting the sensitivity of the sensor to different intensities of light. This behaviour was expected due to the changing offset affecting the luciferase spectrum curve at different light intensities. The offset, using our data, was calculated to be about 0.2V for each dilution. A second graph is shown which takes into account this offset (and removes it), thus showing the presence of blue frequencies. The result was as expected, as the presence of blue frequencies throughout the dilution series is not only detected, but also found to be approximately constant. The raw data of this investigation can be found in the [[Team:Cambridge/Biologger/Labbook|<span style="color:#000066"><u>Lab book.</u></span>]] |
[[File:Norm_light.png|thumb|400px|Normalised sensor data using a dilution series of bioluminescent E.coli- Concentrations on the x-axis are relative therefore an OD 600 value was also taken|center]] | [[File:Norm_light.png|thumb|400px|Normalised sensor data using a dilution series of bioluminescent E.coli- Concentrations on the x-axis are relative therefore an OD 600 value was also taken|center]] | ||
Latest revision as of 01:12, 27 October 2012
Contents |
Judging Form
- Please help the judges by filling out this form. Tell them what medal you think you deserve and why. Tell them which special prizes you should win. Help them find your best parts. Show them how you thought about the safety of your project. Helping the judges will help you too.
- Team: Cambridge
- Region: Europe
- iGEM Year:2012
- Track:Foundational Advance
- Project Name:Parts for a reliable and field ready biosensing platform
- Project Abstract: Implementation of biosensors in real world situations has been made difficult by the unpredictable and non-quantified outputs of existing solutions, as well as a lack of appropriate storage, distribution and utilization systems. This leaves a large gap between a simple, functional sensing mechanism and a fully realised product that can be used in the field.
We aim to bridge this gap at all points by developing a standardised ratiometric luciferase output in a Bacillus chassis. This output can be linked up with prototyped instrumentation and software for obtaining reliable quantified results. Additionally, we have reduced the specialized requirements for the storage and distribution of our bacteria by using Bacillus' sporulation system. To improve the performance of our biosensing platform we have genetically modified Bacillus’ germination speed. Lastly, we demonstrated the robustness of our system by testing it with a new fluoride riboswitch, providing the opportunity to tackle real life problems.
iGEM Medals for non-software teams
- We believe our team deserves the following medal:
- Bronze
- Silver
- √Gold
Because we met the following criteria (check all that apply and provide details where needed)
Requirements for a Bronze Medal
- √Register the team, have a great summer, and plan to have fun at the Regional Jamboree.
- √Successfully complete and submit this iGEM 2012 Judging form.
- √Create and share a Description of the team's project using the iGEM wiki and the team's parts using the Registry of Standard Biological Parts.
- √Plan to present a Poster and Talk at the iGEM Jamboree.
- √Enter information detailing at least one new standard BioBrick Part or Device in the Registry of Standard Biological Parts. Including:
- √Primary nucleaic acid sequence
- √Description of function
- √Authorship
- Safety notes, if relevant.
- √Acknowedgment of sources and references
- √Submit DNA for at least one new BioBrick Part or Device to the Registry.
Additional Requirements for a Silver Medal
- √Demonstrate that at least one new BioBrick Part or Device of your own design and construction works as expected; characterize the operation of your new part/device.
- √Enter this information and other documentation on the part's 'Main Page' section of the Registry
Part Number(s): [http://partsregistry.org/Part:BBa_K911004 BBa_K911004]
Additional Requirements for a Gold Medal: (one OR more)
- Improve an existing BioBrick Part or Device and enter this information back on the Experience Page of the Registry.
Part Number(s): None - √Help another iGEM team by, for example, characterizing a part, debugging a construct, or modeling or simulating their system.
Link to this information on your wiki. Page name: Team:Cambridge/Outreach/Collaboration - √Outline and detail a new approach to an issue of Human Practice in synthetic biology as it relates to your project, such as safety, security, ethics, or ownership, sharing, and innovation.
Link to this information on your wiki.
Page name: Team:Cambridge/HumanPractices/Overview,Team:Cambridge/HumanPractices/MarketResearch,Team:Cambridge/HumanPractices/FutureDirections
iGEM Prizes
All teams are eligible for special prizes at the Jamborees. more... To help the judges, please indicate if you feel you should be evaluated for any of the following special prizes:
- √Best Human Practice Advance
- √Best Experimental Measurement
- Best Model
Please explain briefly why you should receive any of these special prizes:
Best Human Practice Advance:
We feel that we deserve this prize for three reasons:
- We explored the impacts, *both positive and negative*, of synthetic biology as a solution to real world problems, through interviewing professionals working in a relevant field, namely the impact of arsenic water contamination in Bangladesh.
- We recognized existing problems with the way the current direction of synthetic. On going through the registry we found that most of the characterization data for biosensing parts is often neither comparable nor replicable. We have worked to solve this issue, for example with our ratiometric dual channel output.
- *Our project doesn’t stop here*, in Chanel number 6 (Team:Cambridge/HumanPractices/FutureDirections) we considered the future implications and technological applications of our project, as well as the means by which it could be improved by subsequent users. We feel that the end to an iGEM project should not be the conclusion of an idea, but the start of it.
Best BioBrick Measurement Approach:
It is absolutely vital that a quantitative, numerical, robust, and flexible measurement approach exists to relay information to a user that is an accurate representation of the input processed by a biological device. Working from these principles, the following was done:
- We designed and built Biologger, a *cheap, arduino-based, fully functional automatic rotary device* that has an incorporated ratiolumnometer
- Our project is entirely open-sourced and open-platform. We have published source code for the two applications which serve to operate the device, one for PCs and the other for Android devices, as well as the open source circuit design that provides this ratiometric reading. Furthermore, the Android app is able to receive its data wirelessly, which we feel is a great advance in BioBrick measurement.
- Our dual-channel luciferase reporter was successfully tested with a dilution series of E.coli transformed with the Lux Operon (under pBAD) biobrick (Part BBa_K325909) of the Cambridge iGEM 2010 team. It can detect, with good accuracy, both different light intensities, as well as the percentages of blue or orange frequencies in a sample.
- Our device was successfully tested using artificial light to detect different frequencies (colours) as well.
Having done all the above, we believe that this fully open-sourced instrumentation kit (mechanical) chassis, electronics, software code), estimated at *$35.00* (or $85.00 if a Bluetooth modem is required), is a complete BioBrick measurement solution for any and all BioBricks with a light output.
Team_Parts
To help the judges evaluate your parts, please identify 3 of your parts that you feel are best documented and are of the highest quality.
- Best new BioBrick part (natural)
- [http://partsregistry.org/Part:BBa_K911003 BBa_K911003]
- Best new BioBrick part (engineered)
- [http://partsregistry.org/Part:BBa_K911004 BBa_K911004]
- Best improved part(s): None
List any other parts you would like the judges to examine:[http://partsregistry.org/Part:BBa_K911001 BBa_K911001], [http://partsregistry.org/Part:BBa_K911008 BBa_K911009], [http://partsregistry.org/Part:BBa_K911008 BBa_K911008]
Please explain briefly why the judges should examine these other parts:
- Magnesium Sensitive Riboswitch [http://partsregistry.org/Part:BBa_K911001 BBa_K911001]
As a riboswitch sensing construct, this part is an entirely new type of biosensor (along with the fluoride construct) that could potentially change the way we think about designing input genetic circuits. Unlike the fluoride riboswitch, it is a derepression system and therefore serves to demonstrate the principle that riboswitches can be used regardless of whether they turn on or off their reporter. - Fluorescent ratiometric construct for standardizing promoter output [http://partsregistry.org/Part:BBa_K911009 BBa_K911009]
Fluorescence is a major cornerstone for biosensors in the registry, however, most parts do not involve the use of a ratiometric output, which has been shown in the literature to provide much more reliable and meaningful data. This part not only furthers the development of ratiometric measurements in molecular biology but due to the choice of promoters and terminators it can be used to characterize the difference in activity between E. coli and B. Subtilis - Fast Germination (B. subtilis) [http://partsregistry.org/Part:BBa_K911008 BBa_K911008]
This part is entirely novel for the registry and fully utilizes the recombination machinery inherent in the Bacillus chassis. Have spores that can germinate at a faster rate is certainly a worthy achievement and could help with experiments with B. Subtilis that any future iGEM teams may wish to perform.
iGEM Safety
For iGEM 2012 teams are asked to detail how they approached any issues of biological safety associated with their projects.
The iGEM judges expect that you have answered the four safety questions Safety page on your iGEM 2012 wiki.
Please provide the link to that page: Page name: Team:Cambridge/Safety
Attribution and Contributions
For iGEM 2012 the description of each project must clearly attribute work done by the team and distinguish it from work done by others, including the host labs, advisors, and instructors.
Please provide the link to that page, or comments in the box below: Page name: Team:Cambridge/Attributions
Comments
If there is any other information about your project you would like to highlight for the judges, please provide a link to your wiki page here: Team:Cambridge/Overview/DesignProcess
Biologger Results
Having our instrumentation completed, as can be seen in our Instrumentation (Biologger) page, the sensitivity of the sensor placed in the right position was tested using a dilution series of luciferase-producing E.coli. 20ml Cultures were grown overnight from single colonies. The cultures were induced with 40ul of 1.5M arabinose (for a final concentration of 3mM). Cultures were left for 2 1/2 hours for full induction. Subsequently, a culture was pelleted and resuspended in 4ml LB. Doubling dilutions, of volume 2ml, were made from this concentrate, down to 1/8th concentration. 1ml of each 2ml dilution was analysed in each cuvette, which was placed in the cuvette holder we made ourselves. The result was very good. An almost linear relationship was obtained when data were normalised with the sensor value taken in the dark room (the latter set at zero) without using the cuvette holder (1-(sensor value/sensor value in absolute dark)), presenting the sensitivity of the sensor to different intensities of light. This behaviour was expected due to the changing offset affecting the luciferase spectrum curve at different light intensities. The offset, using our data, was calculated to be about 0.2V for each dilution. A second graph is shown which takes into account this offset (and removes it), thus showing the presence of blue frequencies. The result was as expected, as the presence of blue frequencies throughout the dilution series is not only detected, but also found to be approximately constant. The raw data of this investigation can be found in the Lab book.
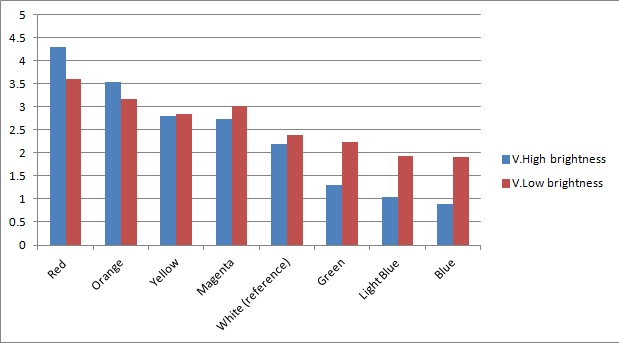
Once the sensor was tested for sensitivity, we tested that our circuitry correctly identified different frequencies (colours) of light. As can be seen below, measurements taken from orange and blue light yield values respectively above and below those from white light (our reference point). The data was taken using constant intensity of light for each case (V.High and V.Low brightness, as specified in the application). This was done with the aid of an Android phone and a specialised software application, called Color Flashlight, downloaded from the official Market.
As expected from the potential-divider design of our circuitry, orange and red frequencies caused the resistance of the LDR with the orange filter to decrease, leading to a higher voltage across the LDR with the blue filter. The opposite effect was observed with blue light. The reason that the white reference point is a bit lower than 2.5V (the expected value for a non-biased circuitry with a 5V source), is because we use resistors of total net resistance 1.67 kΩ before the blue LDR. This was done to bias the circuitry towards blue (i.e. decreasing the starting value, thus the sensor identifies always a bit more blue - this can be shown in previous graphs as well) and thus cause orange light to have a larger impact when present. This was used in an attempt to compensate for the fact that the peak at 560nm (Orange) in MOrange/luciferase fusion spectrum is lower than the one at 490 nm (Blue). Even though we did not manage to test the latter with transformed bacteria, the data collected in all the previous experiments makes us confident that the instrumentation is at least adequately functional.
As the major part of the instrumentation, the bio-electronic interface, had been made and tested, now we turned to testing the other parts of our deveoped kit. This included the mechanical chassis of the prototype, the electronic/mechatronic (sensory and motory) components, and of course the software. The overview of the finished hardware/software can be seen in our Intstrumentation (Biologger) page. Below, the videos showing our instrumentation in action can be seen.
 "
"