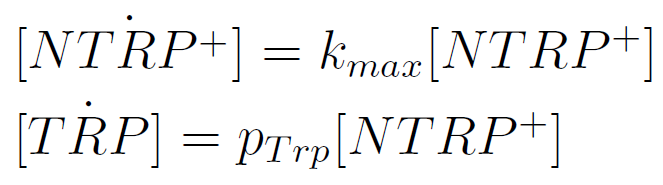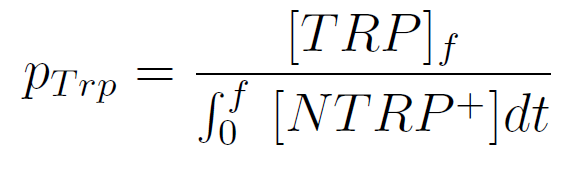Team:Buenos Aires/Results/BBsTesting
From 2012.igem.org

Contents |
After the Jamboree!
When we returned from the Latin America Jamboree we focused all our efforts on completing the neccesary transformations to test our devices and our projet as a whole.
We devided this task in three sections
Week 1&2 : Yeast expression vectors & Transformations
Plasmids
| YFP_TRP+++_TRPZipper | details | |
| YFP_TRP+_TRPZipper | details | |
| YFP_TRP+++_PoliWb | details | |
| YFP_TRP+_PoliWb | details | |
| CFP_HIS+_PoliHa | details | |
| CFP_HIS+_PoliHb | details |
Week 3: synthetic Ecology Characterization
xxoo
BB characterization
xxoo
Secretion Rate of Trp as a function of culture growth
The first step was to actually check if the construct works: do the transformed yeast strains - with the TRP BB - actually secrete TRP into the medium?
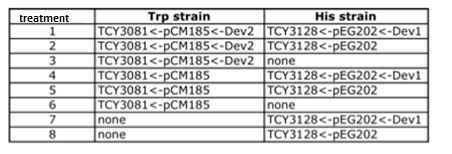
To test this we used the following strains:
- YFP_TRP+++_TRPZipper
- YFP_TRP+++_PolyWb
- YFP_TRP+++
- YFP_TRP+_TRPZipper
- YFP_TRP+_PolyWb
- YFP_TRP+
- YFP
Protocol
- We started 5ml cultures with 3 replica until they reached exponential phase, overnight, using a -T medium.
- Starting OD for the assay 0.1 (exponential phase).
- We measured OD every hour until they reached an OD: 0.8 (5 hs approximately).
- We measured the Trp signal for each culture medium using the spectrofluorometer.
We used a simple model to measure the export rate for all the strains. Since the cultures are in exponential phase, we take
After a few calculations, we find that
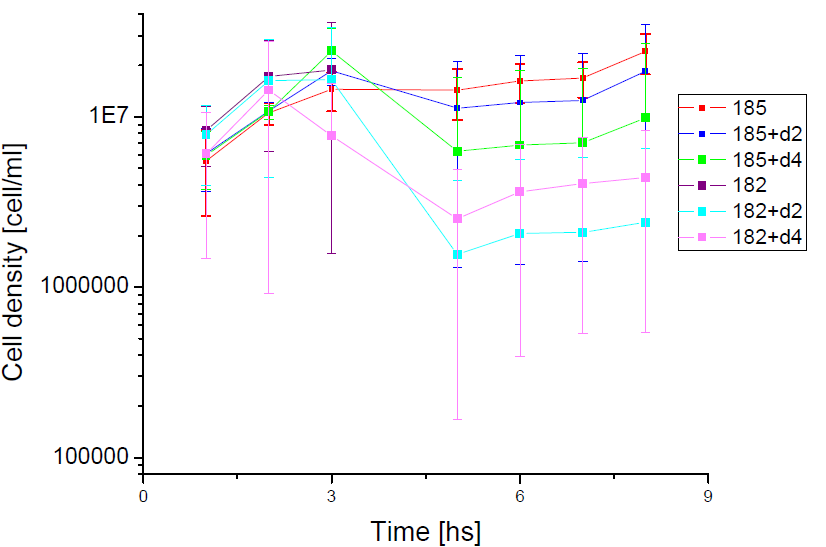
Next we show the average temporal evolution for each strains used,
Figure X. check
Trp Secretion at different His concentrations in medium
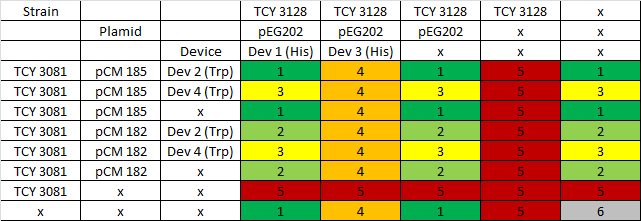
We started cultures in -T of the following strains:
i. strain TCY3081+pCM185 containing device2 ii. strain TCY3081+pCM185 containing device4 iii. strain TCY3081+pCM185 (empty plasmid)
We measured the OD of the cultures after 12 hs. We prepared cultures with increasing concentrations of His in medium (1X, 1/4; 1/8; 0X)and set cultures at initial OD: 0.1. After 5 hours we measured the final OD and Trp in medium.
Strain characterization
Experimental determination of strains death rate
We set out to determine how long can auxotrophic cells[link] survive in media that lacks both Trytophan and Histidine. These values are the death parameters for CFP and YFP strains used in our model[link]. These were taken as equal in the mathematical analysis for simplicity, we'll test whether this approximation is accurate.
Given that our system most likely will present a lag phase until a certain amount of both AmioAcids is accumulated in the media, will the cells be viable until this occurs? This is a neccesary check of our system's feasability.
Protocol
- We set cultures of the two auxotrophic strains without being transformed (YFP and CFP) in medium –HT at an initial OD of 0.01.
- Taking into account that an OD:1 is approximately 3.10^7 cells/ml, we plated 133 µl in order to have around 200 colonies in the first day. Each following day we plated the same amount of µl of the culture and counted the number of colonies obtain in each plate. We set 3 replica of each strain.
 "
"