Team:Bielefeld-Germany/Results/CBD
From 2012.igem.org
Cellulose Binding Domain
In the field of cheap protein-extraction cellulose binding domains (CBD) have made themselfes a name. A lot of publications have been made, concerning a cheap strategy to capture a protein from the cell-lysis with a CBD-tag. Also enhanced segregation with CBD-tagged proteins have been observed. Here the idea is different, we want to take advantage of the binding capacity of binding domains not only for purification reasons (it is still a benefit), but also as an immobilizing-protocol for our laccases.
To make a purification and immobilization-tag out of a protein domain, there are a lot of decisions and characterizations you have to get through.
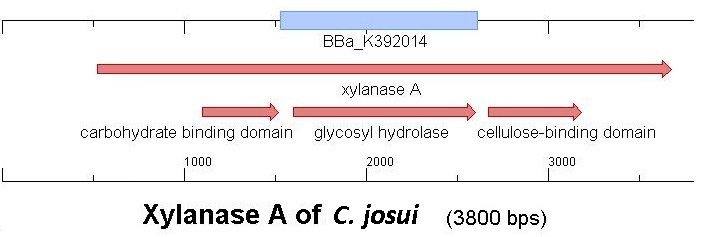
Starting with the choice of the binding domain, the first limitation is accessibility. Our first place to look was, of course the partsregistry. We found a promising Cellulose binding motif of the C. josui Xyn10A gene (<partinfo>BBa_K392014</partinfo>) there and ordered it right from the spot for our project. After some research later concerning the sequence of that BioBrick it turned out that the part is not the CBD of the Xylanase as it should be, but the glycosyl hydrolase domain of the protein (Figure 1). This result made the part useless for our project (complete review) and it was the only binding domain in the partsregistry that fitted to our project.
So we started to search the accessible organisms we had via NCBI for binding-domains, -proteins and -motifs and asked in work-groups if they could help us out. The results of our database research were only two chitin/carbohydate binding moduls within the Bacillus halodurans genome (we ordered that stain for it's laccase <partinfo>BBa_K863020</partinfo>).
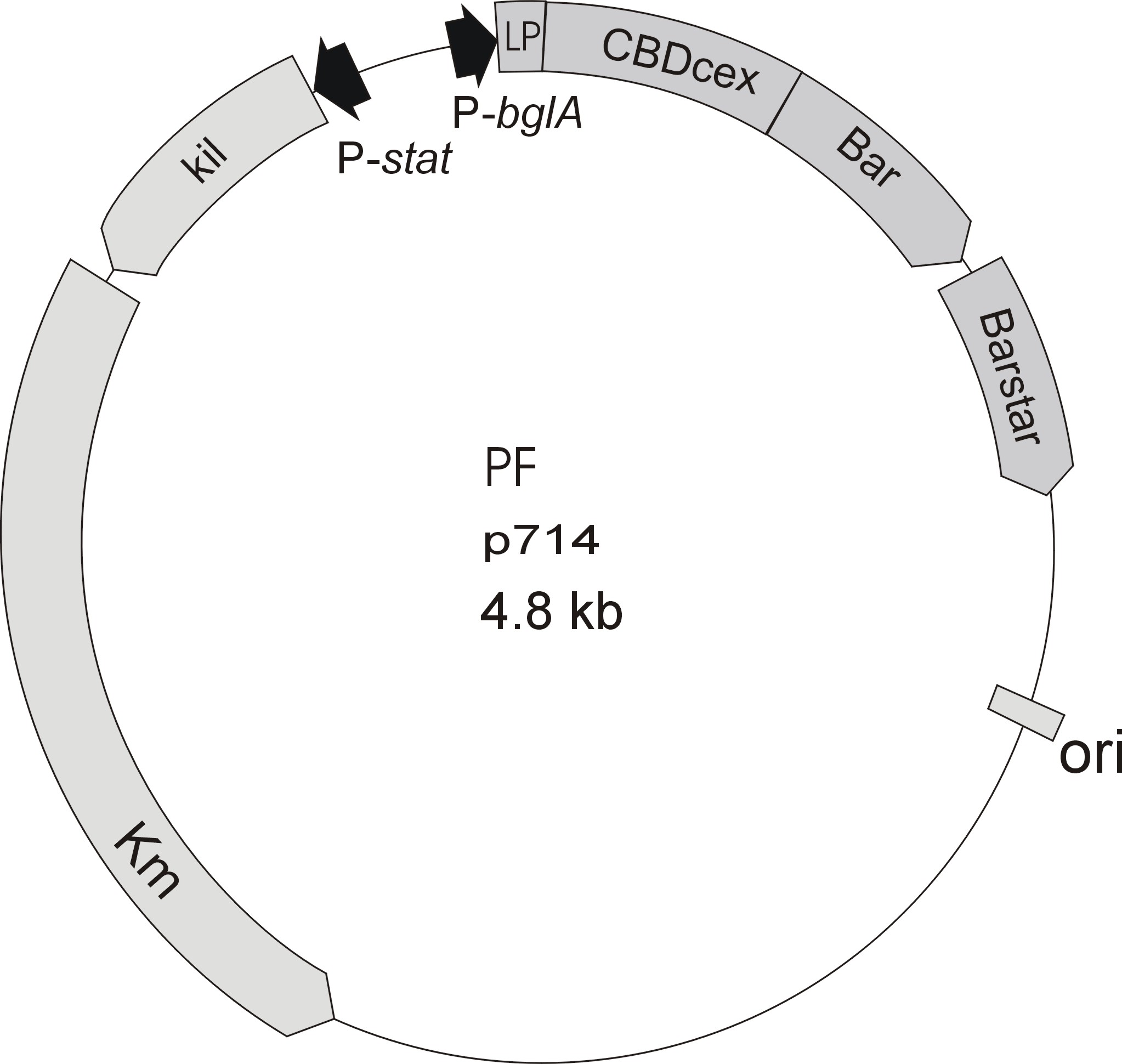
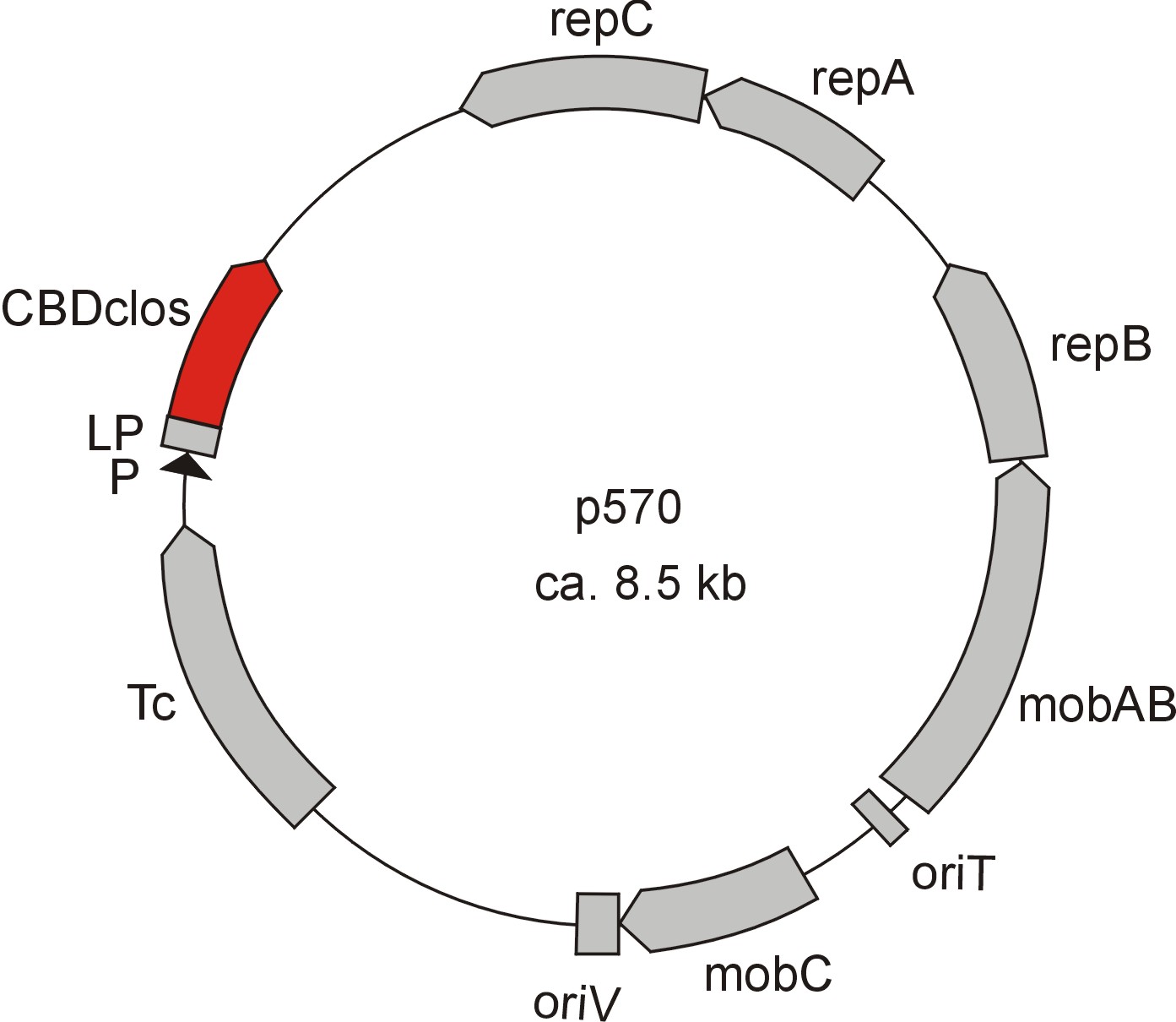
Meanwhile the Fermentation group offered use two plasmids (p570 & p671), containing two different cellulose binding domains. The Cellulose binding domain of the Cellulomonas fimi ATCC 484 exoglucanase gene (CBDcex) and the Cellulose binding domain of Clostridium cellulovorans cellulose binding protein gene (cbp A) (CBDclos). At that point we decided to use these two domains. Staying within the cellulose binding domain-family and leave other protein domains like carbohydrate binding domains aside will keep the results comparable. Like changing to a different binding material would change the binding capacities of both domains in the same way. Also we would stay within bacterial CBD and didn't have to spend time thinking about post-translational modification and glycolization.
To get to know more about these two domains and to get the sequence of the CBDs we consulted NCBI and used the BLAST-tool to identify the cellulose binding domains.

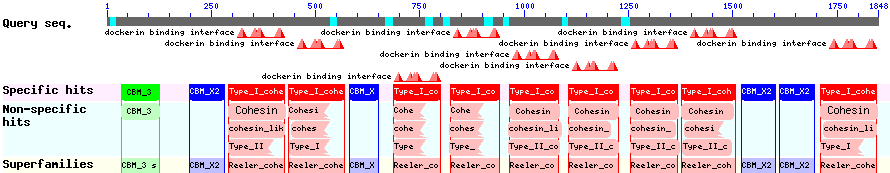
The CBD of the Cellulomonas fimi ATCC 484 exoglucanase gene (Figure 5) is a C-terminal domain of the Cellulose Binding Modul family 2 (pfam00553/cl02709). This means two tryptophan residues are involved in cellulose binding, this type of CBD is only found in bacteria. Also a CBM49 Carbohydrate binding domain is found within the protein domain, where binding studies have showen, that it binds to crystalline cellulose, which could be a possible target for immobilization.
 "
"