Team:Bielefeld-Germany/Results/CBD
From 2012.igem.org
Cellulose Binding Domain
In the field of cheap protein-extraction cellulose binding domains (CBD) have made themselfes a name. A lot of publications have been made, concerning a cheap strategy to capture a protein from the cell-lysis with a CBD-tag. Also enhanced segregation with CBD-tagged proteins have been observed. Here the idea is different, we want to take advantage of the binding capacity of binding domains not only for purification reasons (it is still a benefit), but also as an immobilizing-protocol for our laccases.
To make a purification and immobilization-tag out of a protein domain, there are a lot of decisions and characterizations you have to get through.
Starting with the choice of the binding domain, the first limitation is accessibility. Our first place to look was, of course the partsregistry. We found a promising Cellulose binding motif of the C. josui Xyn10A gene (<partinfo>BBa_K392014</partinfo>) there and ordered it right from the spot for our project. After some research later concerning the sequence of that BioBrick it turned out that the part is not the CBD of the Xylanase as it should be, but the glycosyl hydrolase domain of the protein (Figure 1). This result made the part useless for our project (complete review) and it was the only binding domain in the partsregistry that fitted to our project.
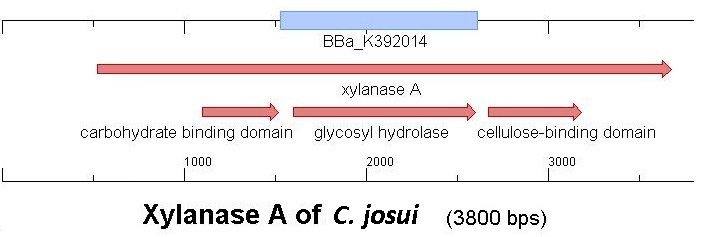
So we started to search the accessible organisms we had for binding domains, proteins and motifs and asked in work-groups if they could help us out.
we decided to take only cellulose binding domains and no protein domains of other carbohydrate binding protein domain families to stay comparable. Else, changing to a different binding material would make a greater difference, than using domains of the same family. "
"