Team:Amsterdam/modeling/generaldesign
From 2012.igem.org
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | {{Team:Amsterdam/stylesheet}} | |
{{Team:Amsterdam/scripts}} | {{Team:Amsterdam/scripts}} | ||
{{Team:Amsterdam/Header}} | {{Team:Amsterdam/Header}} | ||
| Line 24: | Line 24: | ||
== Extending the idea to multiple bits == | == Extending the idea to multiple bits == | ||
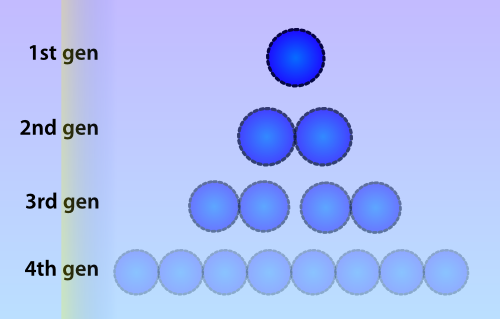
| - | Extending the design idea further, we realized that it should be possible to register and store the presence of multiple signals smultaneously in a single cell. By fusing DNA binding proteins to the methyltransferase and adding their corresponding DNA binding motifs to the bit regions of the DNA. Either traditional bacterial transcriptional regulators could be used for this purpose or the Zinc-Finger Array (ZFA) technology | + | Extending the design idea further, we realized that it should be possible to register and store the presence of multiple signals smultaneously in a single cell. By fusing DNA binding proteins to the methyltransferase and adding their corresponding DNA binding motifs to the bit regions of the DNA. Either traditional bacterial transcriptional regulators could be used for this purpose or the Zinc-Finger Array (ZFA) technology <ref>Fu, Fengli, Jeffry D. Sander, Morgan Maeder, Stacey Thibodeau-Beganny, J. Keith Joung, Drena Dobbs, Leslie Miller, and Daniel F. Voytas. 2009. “Zinc Finger Database (ZiFDB): a repository for information on C2H2 zinc fingers and engineered zinc-finger arrays.” ''Nucleic acids research'' 37 (jan): 279. doi:10.1093/nar/gkn606. [[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2686427\&tool=pmcentrez\&rendertype=abstract|http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2686427\&tool=pmcentrez\&rendertype=abstract]]</ref>. ZFA allows the construction of highly specific artificial protein-DNA interactions (Kaseniit, Perli, and Lu 2011). By fusing the MTase to a Zinc Finger or traditional transcriptional regulator and extending the bit region with the transcriptional regulator’s binding motif, the binding affinity of the fusion protein for the DNA motif could increase about 18-fold (McNamara et al. 2002) (A C5-cytosine MTase was studied here, not in the class of N4C-MTases to which M.ScaI belongs). This will heavily diminish the possible aspecific cross-talk between FPs and bits assigned to other FPs. The same MTase could then be used for different signals with a very low chance of cross-talk between FPs and other signals’ bits. |
Part more specific on read out, unique band lengths | Part more specific on read out, unique band lengths | ||
| Line 145: | Line 145: | ||
== In theory == | == In theory == | ||
<html> | <html> | ||
| - | < | + | |
| - | + | <swf src="https://static.igem.org/mediawiki/2012/f/f4/Timeinferranceanim.swf" width="100%" height="100%" fps="50" caption="Inferring the time of signal onset from the amount of methylated bits" bgcolor="#ff0070"> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<center> | <center> | ||
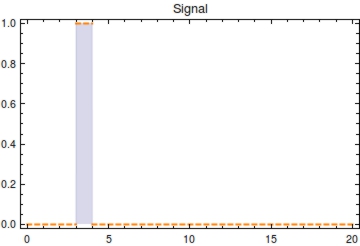
<b>Figure 3</b>. Animation of <math>P_{1}(t)</math> over time with <math>s_{\text{off}}</math> at <math>t = 0</math>. From the measured value of <math>P_{t}</math>, <math>t</math> of measurement can be solved for as is shown using the dashed lines. The value of <math>P_{1}(t)</math> will decrease over time until it becomes 0. An interactive version of this plot has been included in the attached Mathematica file. | <b>Figure 3</b>. Animation of <math>P_{1}(t)</math> over time with <math>s_{\text{off}}</math> at <math>t = 0</math>. From the measured value of <math>P_{t}</math>, <math>t</math> of measurement can be solved for as is shown using the dashed lines. The value of <math>P_{1}(t)</math> will decrease over time until it becomes 0. An interactive version of this plot has been included in the attached Mathematica file. | ||
| Line 176: | Line 170: | ||
== In practice == | == In practice == | ||
| - | + | [[File:bands.jpeg|400px|Gel representations for a range of different <math>F(t)</math> values. Complete methylation of all bits results in a single, bright band at the top of the gel. This indicates the undigested, linearized plasmid. Decreasing the amount of methylated bits shifts the intensity of the top band away to the two bottom bands. These indicate the linearized & successfully digested plasmid]] | |
| - | [[File:bands.jpeg|400px | + | |
| - | + | ||
| - | + | ||
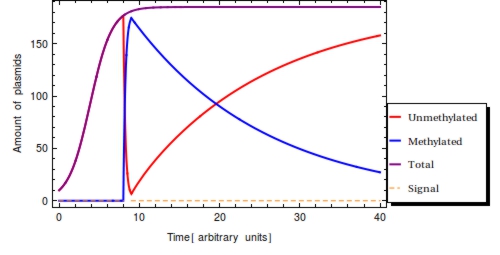
In a typical laboratory situation, doing a restriction enzyme assay on the miniprep-extracted plasmid DNA out of followed by gel electrophoresis will be the most convenient way to assess the methylation status of the bits. The relative intensities of the gel bands can then be used to infer <math>F(t)</math>, which is exemplified in Figure 3. Unmethylated bits will result in successfully digested DNA fragments and thus two bands of shorter DNA fragments. Methylated bits will not be cut and will therefore result in one longer band, shown more to the top of the gel. Thus the top and two bottom gel bands are mutually exclusive as they indicate the same (linearized) plasmid DNA to either be digested, resulting in the two bottom bands, or undigested, resulting in the top band. A high value for <math>P(t)</math> indicates recent detection of the signal, whereas a low value indicates detection to have occurred longer ago. To get a hands-on feel of the effects that the plasmid degradation and replication rate have on <math>F(t)</math>, an interactive version in Mathematica is included in the attached Mathematica file. | In a typical laboratory situation, doing a restriction enzyme assay on the miniprep-extracted plasmid DNA out of followed by gel electrophoresis will be the most convenient way to assess the methylation status of the bits. The relative intensities of the gel bands can then be used to infer <math>F(t)</math>, which is exemplified in Figure 3. Unmethylated bits will result in successfully digested DNA fragments and thus two bands of shorter DNA fragments. Methylated bits will not be cut and will therefore result in one longer band, shown more to the top of the gel. Thus the top and two bottom gel bands are mutually exclusive as they indicate the same (linearized) plasmid DNA to either be digested, resulting in the two bottom bands, or undigested, resulting in the top band. A high value for <math>P(t)</math> indicates recent detection of the signal, whereas a low value indicates detection to have occurred longer ago. To get a hands-on feel of the effects that the plasmid degradation and replication rate have on <math>F(t)</math>, an interactive version in Mathematica is included in the attached Mathematica file. | ||
== References == | == References == | ||
| - | |||
Kaseniit, Kristjan E., Samuel D. Perli, and Timothy K. Lu. 2011. “Designing extensible protein-DNA interactions for synthetic biology.” ''2011 IEEE Biomedical Circuits and Systems Conference (BioCAS)'' (nov): 349–352. doi:10.1109/BioCAS.2011.6107799. [http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=6107799 http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=6107799]. | Kaseniit, Kristjan E., Samuel D. Perli, and Timothy K. Lu. 2011. “Designing extensible protein-DNA interactions for synthetic biology.” ''2011 IEEE Biomedical Circuits and Systems Conference (BioCAS)'' (nov): 349–352. doi:10.1109/BioCAS.2011.6107799. [http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=6107799 http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=6107799]. | ||
| Line 195: | Line 185: | ||
van Steensel, B., and S. Henikoff. 2000. “Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase.” ''Nature biotechnology'' 18 (apr): 424–8. doi:10.1038/74487. [http://www.ncbi.nlm.nih.gov/pubmed/10748524 http://www.ncbi.nlm.nih.gov/pubmed/10748524]. | van Steensel, B., and S. Henikoff. 2000. “Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase.” ''Nature biotechnology'' 18 (apr): 424–8. doi:10.1038/74487. [http://www.ncbi.nlm.nih.gov/pubmed/10748524 http://www.ncbi.nlm.nih.gov/pubmed/10748524]. | ||
| - | |||
</div> | </div> | ||
Revision as of 10:14, 23 September 2012
 "
"