Team:Amsterdam/achievements/stochastic model
From 2012.igem.org
(Difference between revisions)
(→Comparison of SSA implementations) |
(→Introduction) |
||
| Line 19: | Line 19: | ||
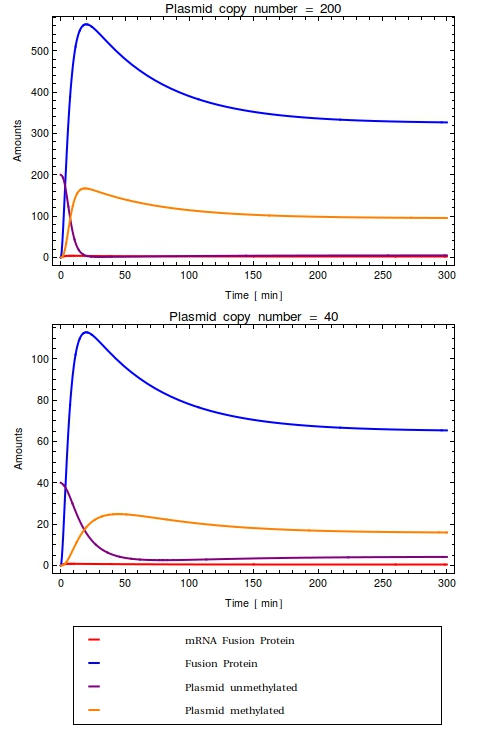
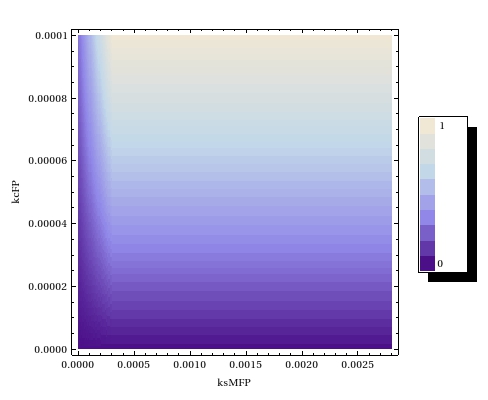
Before starting the experiments, a lot of crucial parameters on the molecular system were still unknown. Both the leaky expression rate of the operon and the activity of the fusion protein were hard to model because of this. Sensible predictions on how the molecular system we were designing was going to function exactly therefore seemed impossible. We already expected the promoter/sensor we were going to test, pLac, to show some leaky expression. As to how much background activity this was going to cause, we couldn't predict and thus we proceeded to test out the construct in the lab. The [[Team:Amsterdam/Lab|laboratory results]] seemed to suffer from two problems: | Before starting the experiments, a lot of crucial parameters on the molecular system were still unknown. Both the leaky expression rate of the operon and the activity of the fusion protein were hard to model because of this. Sensible predictions on how the molecular system we were designing was going to function exactly therefore seemed impossible. We already expected the promoter/sensor we were going to test, pLac, to show some leaky expression. As to how much background activity this was going to cause, we couldn't predict and thus we proceeded to test out the construct in the lab. The [[Team:Amsterdam/Lab|laboratory results]] seemed to suffer from two problems: | ||
| - | * A basal methylation activity that was always recorded even in absence of the signal | + | * A basal methylation activity that was always recorded even in absence of the signal.This background noise reduces the dynamic range of our system; there is no way to tell whether observed band intensities are due to the signal having passed the ''Cellular Logbook'' [[Team:Amsterdam/modelling/timeinferrance|some time ago]] or simply because of the background noise. We conclude that storing a signal using the ''Cellular Logbook'' requires a transcriptional regulator that has a high repressing quality in order to diminish the background activity. |
| - | This background noise reduces the dynamic range of our system; | + | * A lack of a fully induced state for the pLac operon, but not for the later tested pBAD operon. We suspect this problem to be caused by a low uptake rate of the signalling compound (lac operon inducer IPTG) compared to the amount of repressor LacI present in the system controlling the operons. Especially for the part characterization experiments in which the pLac-MTase construct was cloned into a high copy number plasmid, this could be a problem. This problem is solved by adding the ''LacY''-gene that codes for permease to the existing construct. Inclusion of this gene might also lead to higher background methylation percentages in the negative controls, however. |
| - | there is no way to tell whether observed band intensities are due to the signal having passed the ''Cellular Logbook'' [[Team:Amsterdam/modelling/timeinferrance|some time ago]] or simply because of the background noise. | + | |
| - | We conclude that storing a signal using the ''Cellular Logbook'' requires a transcriptional regulator that has a high repressing quality in order to diminish the background activity. | + | |
| - | * A lack of a fully induced state for the pLac operon, but not for the later tested pBAD operon. | + | |
| - | We suspect this problem to be caused by a low uptake rate of the signalling compound (lac operon inducer IPTG) compared to the amount of repressor LacI present in the system controlling the operons. | + | |
| - | Especially for the part characterization experiments in which the pLac-MTase construct was cloned into a high copy number plasmid, this could be a problem. | + | |
| - | This problem is solved by adding the ''LacY''-gene that codes for permease to the existing construct. | + | |
| - | Inclusion of this gene might also lead to higher background methylation percentages in the negative controls, however. | + | |
== Stochastic Differential Equations == | == Stochastic Differential Equations == | ||
Revision as of 20:25, 26 September 2012
 "
"