Team:Amsterdam/achievements/stochastic model
From 2012.igem.org
(Difference between revisions)
(→Comparison of SSA implementations) |
|||
| Line 44: | Line 44: | ||
Additionally, we've also extended StochPy by writing a supplementary tool [Team:Amsterdam/Software/MDL2LateX MDL2LaTeX], that eases the publishing of models developed with StochPy. | Additionally, we've also extended StochPy by writing a supplementary tool [Team:Amsterdam/Software/MDL2LateX MDL2LaTeX], that eases the publishing of models developed with StochPy. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=Leakiness and background noise= | =Leakiness and background noise= | ||
| Line 84: | Line 59: | ||
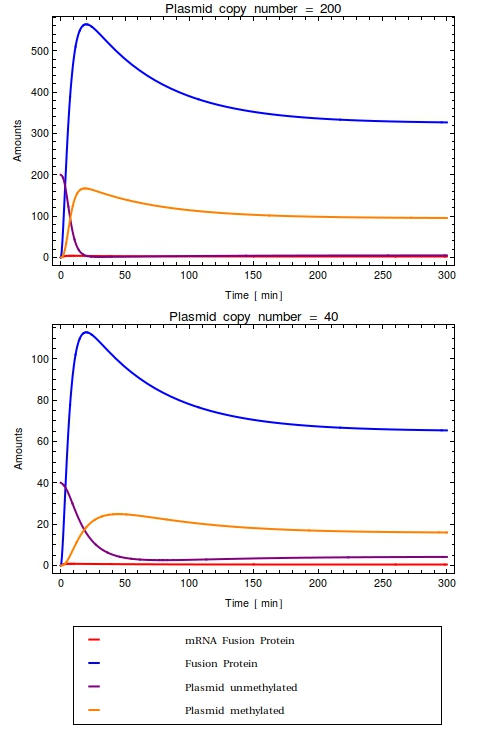
In order to not exclude the possible qualitative difference in model behaviour when all molecular reactions are computed individually, the system has also been modelled stochastically. To make the two systems perfectly comparable, the used rates are qualitatively identical to the ones used in the ODE system. | In order to not exclude the possible qualitative difference in model behaviour when all molecular reactions are computed individually, the system has also been modelled stochastically. To make the two systems perfectly comparable, the used rates are qualitatively identical to the ones used in the ODE system. | ||
Furthermore, the same parameter values have been used. | Furthermore, the same parameter values have been used. | ||
| - | |||
| - | |||
==ODE Model definition== | ==ODE Model definition== | ||
| Line 108: | Line 81: | ||
\text{O} = \text{PlasU} + \text{PlasM} | \text{O} = \text{PlasU} + \text{PlasM} | ||
$$ | $$ | ||
| + | |||
| + | == Leaky expression rate == | ||
| + | There are discrepancies between the definitions used in the literature on what leaky expression exactly is: | ||
| + | * The most favoured definition seems to be the that leaky operons show low expression rates even when bound by a negative transcriptional regulator. In this perspective the transcriptional regulator is regarded to be unable to completely silence gene expression. In this case the only way to get a value for leaky expression is to measure it experimentally. In this process, care should be taken to be specific about what version of the operon exactly is being used. Some repressors are known to regulate transcription by binding to DNA regions that are very distant from the controlled operon. 'Plasmid versions' of these operons might not contain these distantly located binding sites and are therefore less likely to be optimally repressed. | ||
| + | * Another definition is that the TF ''can'' fully silence gene expression and that leaky expression is caused by seldomly occurring dissociation events between between the TF and the DNA operator sequence. | ||
| + | During these short time windows of opportunity, the RNA polymerases would then be able to quickly transcribe a small amount of RNA. | ||
| + | Multiplying the maximal rate of transcription by the time fraction during which the polymerase is able to bind gives an indication of the leaky transcription rate defined this way. | ||
| + | |||
| + | The following relationship must always be true: $[O_{\text{Total}}] = [O_{\text{Free}}] + [R_{2}O]$ with $R_{2}O$ denoting the operon bound by a dimerized repressor (as LacI, the repressor of the Lac operon, functions). $R_{2}O$ is defined as $\frac{[R_{2}][O]}{K_{d}}$. Solving for $O_\text{Free}$: | ||
| + | |||
| + | $$ | ||
| + | [O_{\text{Free}}] = [O_{\text{Total}}] (1 + \frac{[R_{2}]}{K_{d}}) | ||
| + | $$ | ||
| + | |||
| + | We immediately see that the $[O_{\text{Free}}]$ depends on $[R_{2}]$ and $O_{\text{Total}}$. | ||
| + | The dissociation constant of the dimerized LacI-repressor for its operator sequence has been determined to be $10^{-12} M $ and the volume of ''E. coli'' is $0.7\ \mu m^{3}$. | ||
| + | The transcription rate in E.coli is 40-80 bp/s. | ||
| + | |||
| + | More simply, the leaky expression rate can also be retrieved from this [[http://oregonstate.edu/instruction/bb492/lectures/Regulation.html|website]], which states a 1000-fold decrease in expression of the repressor-bound operon compared to the free operon. | ||
| + | |||
| + | == Fusion protein catalysis rate == | ||
| + | The catalysis rate $k_{cat}$ has not been determined for the MTase that we are using, M.ScaI. | ||
| + | However, for another MTase with the same [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.1.1.113 EC2.1.1.113 number] as M.ScaI, BamH1, the $k_{cat}$ has been determined to be $0.0175$ ([[#Cheng|1]]). | ||
| + | |||
| + | == Cell growth rate == | ||
| + | Our own experiments showed that the bacterial strain <math>DH5\alpha</math> transformed with two of our preliminary constructs had growth rates (<math>\mu</math>) between $80\ min^{-1}$ and $90\ min^{-1}$ ([Team:Amsterdam/Project/Growth_curves Experimental growth curves]). | ||
| + | In these simulations, cellular division was assumed to be either constant at 80 for single cells models or Gaussian distributed with $\mu = 80$ and $\sigma = 2$ in the cell division model. | ||
| + | |||
<table align="right"> | <table align="right"> | ||
<tr><th>Parameter</th><th>Value</th></tr> | <tr><th>Parameter</th><th>Value</th></tr> | ||
Revision as of 20:22, 26 September 2012
 "
"