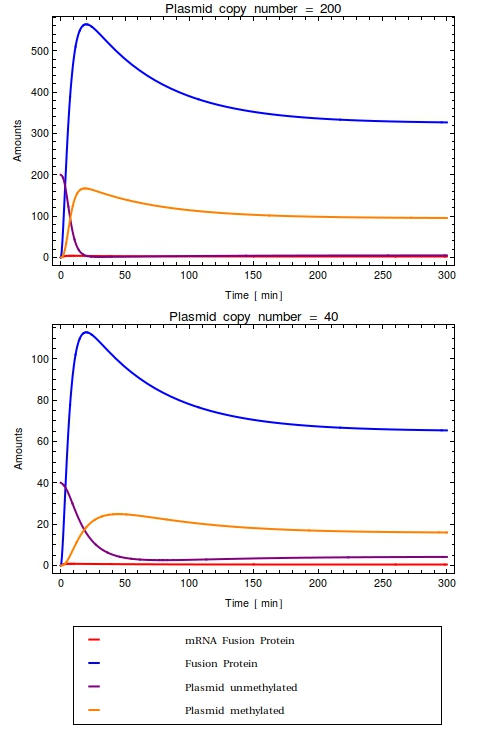
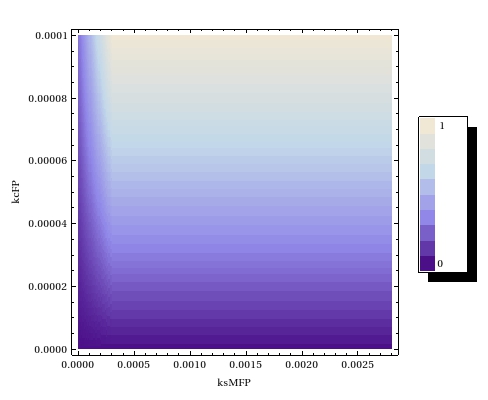
Team:Amsterdam/achievements/stochastic model
From 2012.igem.org
(Difference between revisions)
(→ODE Model definition) |
|||
| Line 13: | Line 13: | ||
* the leaky transcription rate of the mRNA for the fusion protein (FP), denoted by $k_{c\text{MFP}}$ | * the leaky transcription rate of the mRNA for the fusion protein (FP), denoted by $k_{c\text{MFP}}$ | ||
* the catalysis rate of the FP, or the rate with which it methylates umethylated plasmids | * the catalysis rate of the FP, or the rate with which it methylates umethylated plasmids | ||
| + | |||
== Leaky expression rate == | == Leaky expression rate == | ||
| + | |||
There are discrepancies between the definitions used in the literature on what leaky expression exactly is: | There are discrepancies between the definitions used in the literature on what leaky expression exactly is: | ||
* The most favoured definition seems to be the that leaky operons show low expression rates even when bound by a negative transcriptional regulator (REF). In this perspective the transcriptional regulator is regarded to be unable to completely silence gene expression. In this case the only way to get a value for leaky expression is to measure it experimentally. In this process, care should be taken to be specific about what version of the operon exactly is being used. Some repressors are known to regulate transcription by binding to DNA regions that are very distant from the controlled operon. 'Plasmid versions' of these operons might not contain these distantly located binding sites and are therefore less likely to be optimally repressed. | * The most favoured definition seems to be the that leaky operons show low expression rates even when bound by a negative transcriptional regulator (REF). In this perspective the transcriptional regulator is regarded to be unable to completely silence gene expression. In this case the only way to get a value for leaky expression is to measure it experimentally. In this process, care should be taken to be specific about what version of the operon exactly is being used. Some repressors are known to regulate transcription by binding to DNA regions that are very distant from the controlled operon. 'Plasmid versions' of these operons might not contain these distantly located binding sites and are therefore less likely to be optimally repressed. | ||
Revision as of 17:40, 26 September 2012
 "
"