Refactored Decaffeination Operon
From 2012.igem.org
Erik.quandt (Talk | contribs) (→Experiment 2: Constructon of a refactored decaffeination operon) |
Erik.quandt (Talk | contribs) (→Experiment 2: Constructon of a refactored decaffeination operon) |
||
| Line 1: | Line 1: | ||
| + | {{Template:Austin_Texas/Stylesheet}} | ||
| + | |||
| + | <html> | ||
| + | <ul class="cssmenu"> | ||
| + | <li class="home"><a href="/Team:Austin_Texas" title="home"><span class="displace">Home</span></a></li> | ||
| + | <li class="team"><a href="/Team:Austin_Texas/Team" title="team"><span class="displace">Team</span></a></li> | ||
| + | <li class="official_team_profile"><a href="https://igem.org/Team.cgi?year=2012&team_name=Austin_Texas" title="official_team_profile"><span class="displace">Official Team Profile</span></a></li> | ||
| + | <li class="project"><a href="/Team:Austin_Texas/Project" title="project"><span class="displace">Project</span></a></li> | ||
| + | <li class="parts_submitted"><a href="/Team:Austin_Texas/Parts" title="parts_submitted"><span class="displace">Parts Submitted</span></a></li> | ||
| + | <li class="modelling"><a href="/Team:Austin_Texas/Modeling" title="modeling"><span class="displace">Modeling</span></a></li> | ||
| + | <li class="notebook"><a href="/Team:Austin_Texas/Notebook" class="selected" title="notebook"><span class="displace">Notebook</span></a></li> | ||
| + | <li class="safety"><a href="/Team:Austin_Texas/Safety" title="safety"><span class="displace">Safety</span></a></li> | ||
| + | <li class="attributions"><a href="/Team:Austin_Texas/Attributions" title="attributions"><span class="displace">Attributions</span></a></li> | ||
| + | </ul> | ||
| + | </html> | ||
| + | |||
| + | |||
==Experiment 2: Constructon of a refactored decaffeination operon== | ==Experiment 2: Constructon of a refactored decaffeination operon== | ||
Revision as of 18:35, 20 August 2012
Experiment 2: Constructon of a refactored decaffeination operon
2a. Phusion PCR
Primers:
EQ_pSB1c3_NdmA_for: TAGGTACAGTGCTAGCTACTAGAGAAATCAAATTAAGGAGGTAAGATAAATGGAACAGGCAATCATTAATGATGAACGGG
EQ_pBSC1C_NdmA_2: TGATTTCTGGAATTCGCGGCCGCTTCTAGAGATTAAGGAGGTAAGATAAATGGAACAGGCAATCATTAATGATGAACGGG
EQ_pSB1C3_Gib_rev: TTGATTTCTCTAGTAGCTAGCACTGTACCTAGGACTG
EQ_NdmA_rev: TTATATGTAGCTCCTATCGCTTTCAATGACTGGG
EQ_RBS_NdmB_for: gtCATTGAAAGCGATAGGAGCTACATATAATCTAGAGAAAGAGGAGAAATACTAGATGAAAGAACAGCTCAAGCCGCTG
EQ_NdmB_rev: ccggtctcgcTTACTGTTCTTCTTCAATAACATTCGTCAAGACG
EQ_RBS_NdmC_for: GAAGAAGAACAGTAAgcgagaccggTCTAGAGAAAGAGGAGAAATACTAGATGTCTACTGACCAAGTAATTTTTAACGactgg
EQ_NdmC_rev: TTAGTCCCGCAGAGCACCATATTGCac
EQ_RBS_NdmD for: GCAATATGGTGCTCTGCGGGACTAATCTAGAGAAAGAGGAGAAATACTAGATGAACAAACTTGACGTCAACCagtgg
EQ_NdmD_pBS1C_rev: TTATACAGCTCGTCCATACCGTGGGTGATGCCCGGCCTCACAGATCGAGAACGATT
EQ_pBS1C_Gib_for: GGGCATCACCCACGGTATGGACGAG
EQ_pBS1C_rev_2: CTCTAGAAGCGGCCGCGAATTCCAGAAATCA
10 uL 5x Phusion HF Buffer
1 uL 10mM dNTPs
5 uL 5uM forward primer
5 uL 5uM reverse primer
1 uL DMSO
1 uL template
26.5 uL H20
--
.5 uL phusion polymerase
rxns:
Vector PCRs
1. Bba_k515105 + EQ_pSB1C3_Gib_for + EQ_pSB1c3_gib_rev
2. Bba_k515105 + EQ_pSB1C3_Gib_for + EQ_pSB1c3_gib_rev_2
Insert PCRs
3. CBB5 cells + EQ_NdmA_for + EQ_NdmA_rev
4. CBB5 cells + EQ_pSB1C3_NdmA_2 + EQ_NdmA_rev
5. CBB5 cells + EQ_RBS_NdmB_for + EQ_NdmB_rev
6. CBB5 cells + EQ_RBS_NdmC_for + EQ_NdmC_rev
7. CBB5 cells + EQ_RBS_NdmD_for + EQ_NdmD_pSB1C_rev
PCR cycling:
98 deg x 3 min
--
98 deg x 30s
58 deg x 20s x30 cycles
72 deg x 2 min
--
72 deg x 10 min
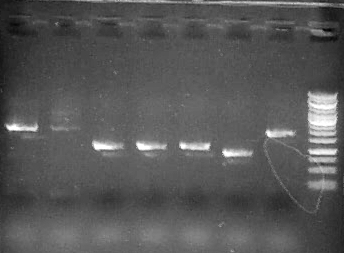
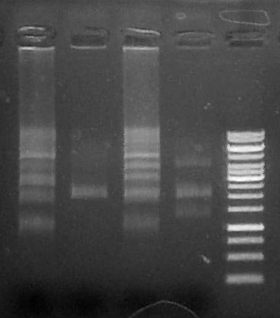
Run 1 uL 2a1-7, 3.5 uL Fermentas 1kb Generuler ladder
2b. PCR purification
Qiagen PCR purification protocol
Elute in 35 uL h20
nanodrop concentrations:
1. vector 1: 289.0 ng/uL
2. vector 2: 85.6 ng/uL
3. NdmA_1: 139.8 ng/uL
4. NdmA_2: 144.5 ngl/uL
5. NdmB: 144.6ng/uL
6. NdmC: 146.1 ng/uL
7. NdmD: 104.8ng/uL
2c: DpnI digest of vector PCRs
1.
15 uL vector 1
5 uL 10x NEB 4 Buffer
1 uL dpnI
29 uL H20
--
50 uL
2.
30 uL vector 2
5 uL 10x NEB 4 Buffer
1 uL dpnI
14 uL H20
--
50 uL
37 deg x 2 hrs
2e. Purification
Standard Qiagen PCR purification protocol. Elute in H20.
2F. Gibson cloning
pmols = weight(ng) x 1000 / (bp x 650 daltons)
Will use .05 pmol of vector, .1 pmol for each insert
Vec 1 (.05pmol) = need 68.57ng / (79.5ng/uL) = .86uL
Vec 2 (.05pmol) = need 68.57ng / (132.5ng/uL) = .52uL
NdmA1 (.1pmol) = need 71.5ng / (139.8ng/uL) = .51 uL
NdmA2 (.1pmol) = need 71.5ng / (144.5ng/uL) = .49 uL
NdmB (.1pmol) = need 73.4ng / (144.6ng/uL) = .50 uL
NdmC (.1pmol) = need 58.8ng / (146.1ng/uL) = .40 uL
NdmD (.1pmol) = need 120.ng / (104.8ng/uL) = 1.15 uL
rxns:
1.
.86 uL vec 1
.51 uL ndmA_1
.50 uL NdmB
.40 uL NdmC
1.15 uL NdmD
15 ul 1.33x Gibson Master Mix
1.58 uL H20
--
20 uL total
2.
.86 uL vec 1
15 uL 1.33x Gibson Master Mix
4.14 uL H20
--
20 uL total
3.
.52 uL Vec 2
.51 uL ndmA_1
.50 uL NdmB
.40 uL NdmC
1.15 uL NdmD
15 ul 1.33x Gibson Master Mix
1.94 uL H20
--
20 uL total
4.
.52 uL vec 1
15 uL 1.33x Gibson Master Mix
4.14 uL H20
--
20 uL total
50 deg thermocycler x 60 minutes
Run 5 uL each reaction (1-4)on .8% agarose gel + 3.5 ul Fermentas 1kb GeneRuler
Desalted gibson reactions on .025uM Nitrocellulose membranes x 20 minutes
2g. Electroporation
Decided to not proceed with promoterless construct (2F3,4)
1 uL desalted gibson reaction (2F1,2) ->50 uL electrocompetent BW25113-GuaB -> 1mL SOC -> 1hr incubation @ 37 deg
2j. Selective growth in Caffeine/Theophylline
dilute 60 uL 2g transformations (1-2) -> 3mL Mineral M9 media + 34ug/mL chloramphenicol +/- 100mg Caffeine or Theophylline. Place on 30 deg shaker x 48 hrs
Results (growth):
2g1 (+operon) 2g2 (vector only) 1. M9 - - 2. M9 + caffeine - - 3. M9 + Theophylline + _
Result: construct enables growth (demethylation) on Theophylline. NdmC is not fuctional.
Streak 2j1 theophylline enriched culture onto LB-chloramphenicol plate for isolation of single colonies.
2K Plasmid Prep
Pick 2 colonies from theophylline enriched streak for plasmid prep. Prep 5mL culture.
2L Restriction Digest
Perform restriction digest to analyze insert size of plasmids
1 uL 2k1,2
1 uL 10x NEB 3
.5 uL NotI
6.5 uL H20
--
10 uL
37 deg x 1hr
Run 5 uL on .8% agarose gel
insert gel image (on darkroom computer?)
Result: both clones show anticipated ~5kB band corresponding to the complete operon.
 "
"