Team:WashU/mobile/Week5
From 2012.igem.org
YLC
At the end of last week we ran a gel of the digests from the fluorescent proteins and extracted DNA from the gels. At the beginning of this week, we used the nanodrop to see how much DNA we had retrieved from the gel. The nanodrop revealed barely imperceptible maxima to indicate that we had almost undetectable amounts of DNA by the nanodrop. In fairness, the bands were tiny to begin with, leading to our poor results. Our mentors assured us that we probably had enough DNA for a ligation, so we plan to run that once the promoter digest works.
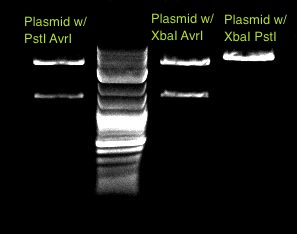
We accidentally grew up the new promoter, J23119, in both plain LB and LB + amp, yet still had colonies in both media, and thus decided to run two digests to ascertain that we have our promoter in both cultures. We used the biobrick protocol with a slight modification - we used NEBuffer 4 instead of NEBuffer 2, since the enzymes we were cutting with, E and S, also have 100% activity in NEBuffer 4 - and ran a gel to ensure that we had pure promoter. The gel is shown below:

Unfortunately, the digest did not seem to work. We attempted to digest the same DNA once more, using NEBuffer 2 instead of NEBuffer 4, and ran a second gel. This gel also reveals that the digest was unsuccessful, as we see only one band when we expect to find two bands. We believe that our enzyme SpeI has not been cutting properly.

To test what was going wrong, we decided to run one final gel. We digested our promoter, just like before, but used four different combinations of enzymes. We digested with EcoRI and SpeI (ES), XbaI and SpeI (XS), EcoRI and PstI (EP), and XbaI and PstI(XP). This gel was also a failure, as we only saw one band for each well, when we should have seen two.

We have also decided to use PCR to amplify our DNA, as we are getting extremely low yields from our gel extractions. Thus, we designed the primers today.
Tuesday, June 26
YLC/Saffron in a Kan
We decided to figure out what was going wrong with our digests by running two new gels, one for the E. coli part of the project and one for the YLC project. First, we used the nanodrop to measure how much DNA we had for each of our samples. The data is shown below.
| Sample | nanograms/microliter |
|---|---|
| 1: YFP | 83.0 |
| 2: GFP | 121.6 |
| 3: mRFP | 63.8 |
| 4 - I: eCFP | 83.9 |
| 4 - II: eCFP | 99.4 |
| 5: promoter BBa_J23100 | 127.0 |
| 8: mCherry | 183.4 |
| 9: carotenoids inE. coli | 84.1 |
| 10: crtZ inE. coli | 37.5 |
| Promoter J23119 | 34.5 |
Then, we ran digests of 9, 10, 5 and BBa J22119 using controls, DNA cut with the enzymes as stated in the biobrick assembly protocol(shown with a "c" below), and then DNA cut with enzymes and then treated with phosphatase (shown as P- on the gel). For the YLC project, we ran 1, 2, 3, 4 and 8. [Gels shown below]


Our digests appear to be successful this time. We extracted the DNA for all of the YLC runs except for 4 and also extracted 9 from the first gel, and then purified all of the above bands.
YLC
We finally ordered the primers from Sigma today and they will arrive in a few days.
Wednesday, June 27
YLC/Saffron in a Kan
After our meeting with our mentor Dr. Dantas this morning, we decided to try several ligations. We agreed to proceed with the J23119 promoter for the future constructs.
One ligation was considered to insert the RBS-ORF-TERM into the J23119 plasmid directly without cutting out the J23119 promoter and proceeding with the usual triple ligation. J23119 in its pSB1A2 plasmid was cut with SpeI and PstI to try to open up the plasmid for the insertion of the RBS-ORF-TERM constructs behind the promoter in a double ligation rather than triple ligation. The main problem with this ligation was that both pieces used came from pSB1A2 plasmid backbones carry Amp resistance so the ability to select was greatly reduced. However, the use of only two substrates of comparable size in the ligation was more favorable than the triple ligations with sizes of ~1 kb, ~35 bp, ~2.5 kb. So there was a win-lose element that we wanted to try out.
The other ligation was traditional triple ligation using gel purified samples of the ORF-region. The plasmid and promoter were both not gel purified. We attempted the ligation knowing that it was not very likely given that the NanoDrop results were poor for the post-purification DNA. (This DNA was recovered from a diagnostic gel that looked good so was thus cut. Even though it was not a fully adequate amount of DNA to cut out, we were hopefully and went for it.) Below is a table of our NanoDrop results:
 "
"