Team:Potsdam Bioware/Lab/Labjournal/September
From 2012.igem.org
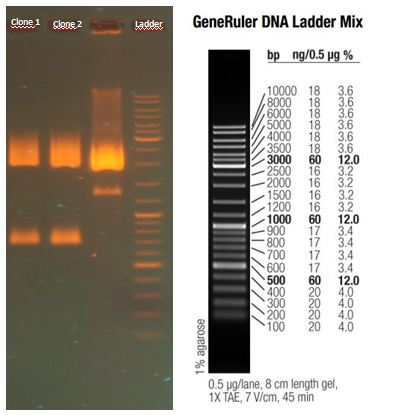
AID
2012-09-03
Miniprep of mVENUS construct and wildtype AID from transfected CHO cells
Investigators:
Rico
Aim:
- Miniprep of YFP-construct and wildtype AID from transfected CHO cells to proof whether it is possible to transform E.coli with the purified construct for sequencing
Method:
- Miniprep of cells
- Elution with 50 µL
Results:
CHO transfected with YFP WT-AID = 2.8 ng/µL
Transformation of mVenus and wildtype AID isolated samples and pCEP4
Investigators: Tom
Time: 2012-09-03
Materials:
- Bunsen burner, Agar plates with chloramphenicol
- icebox
- competent E. coli cells (XL 1 Blue)
- Venus plasmid - sample 1
Method:
Transformation via manual, 10 µl of miniprep sample and 1 µL of pCEP4 were used
Plate incubation start: 1:30 pm
Results:
ready for growing mutants to pick clones
Further tasks:
picking clones
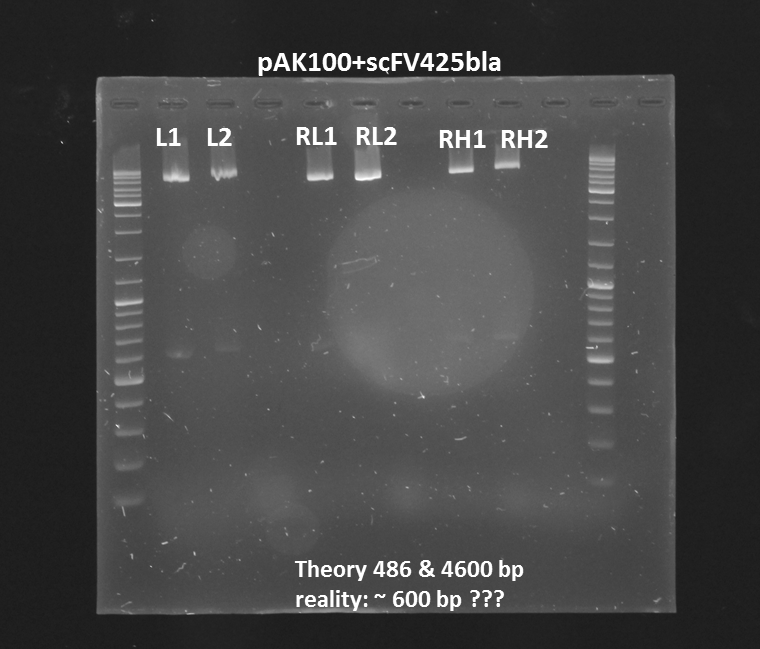
Test digestion (Pst1) & gel electrophoresis of pAK100+scFV
Investigators:
Chris, Rico
Aim:
*testing the miniprep samples of pAK100+scFV
Results:
a 600 bp instead of 480bp
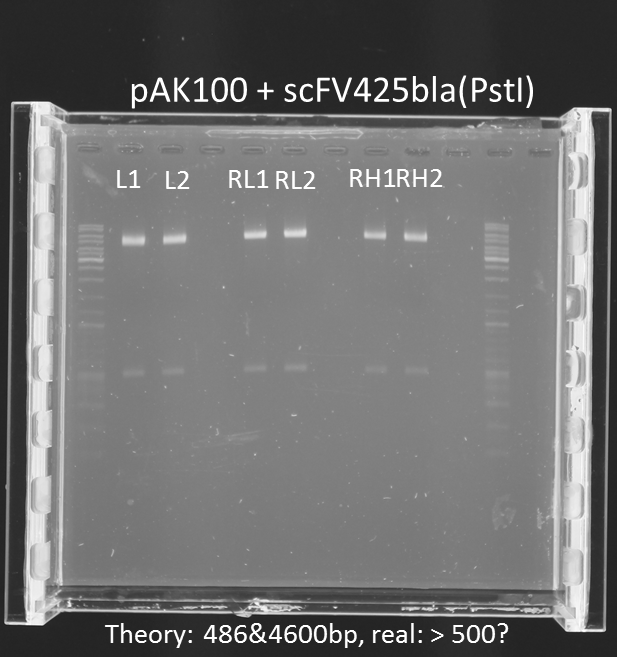
repetition of test digestion (Pst1) & gel electrophoresis of pAK100+scFV
Investigators:
Chris
Aim:
*testing the miniprep samples of pAK100+scFV
Results:
a 600 bp instead of 480bp
2012-09-04
preparation of competent E.coli XL1 blue cells
Investigators:
Basia, Chris
Method:
via standard manual
Results:
50 Eppendorf tubes with 100 µL competent XL1 blue
further tasks:
testing competence
Transfection of CHO cells with wt AID, AID without NES, with NLS+Kozak sequence, AID without NES, with NLS+Kozak sequence+eGFP
Investigators:
Rico
Aim:
- Transfection of CHO-cells with wt AID + small antibody construct, AID without NES, with NLS+Kozak sequence + small antibody construct, AID without NES, with NLS+Kozak sequence+eGFP + small antibody construct, small antibody construct
Method:
- 7 µg total DNA was used for transfection in 700 µL total volume:
- 3.5 µg wt AID + 3.5 µg small antibody construct
- 3.5 µg AID without NES, with NLS+Kozak sequence + 3.5 µg small antibody construct
- 3.5 µg AID without NES, with NLS+Kozak sequence+eGFP + 3.5 µg small antibody construct
- 7 µg small antibody construct
- 17.5 µL PEI in 700 µL Optimum/PEI-Mix
- vortex for 10 s 3 x
- 2 min incubation
- mix DNA solution with PEI solution (1:1)
- 15 min incubation
- add 400 µL PEI/DNA-solution to the CHO cells
send the DNA to sequencing
Investigators:
Tom S., Chris
| sample | GATC number | Seq. Primer |
| CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA 2-1-2 | II3637 | pSB1C3 Forward |
| CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA 2-1-2 | II3638 | pSB1C3 Reverse |
| CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA 2-1-2 | II3639 | AID-C-Term Forward |
| CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA 2-1-2 | II3640 | eGFP-N-Term Reverse |
| Pak100+ScFv L1 | II3688 | GATC_std_RPC |
| Pak100+ScFv L1 | II3689 | GATC_std_sr_Hind3-672953 |
| Pak100+ScFv RH1 | II3690 | GATC_std_RPC |
| Pak100+ScFv RH1 | II3691 | GATC_std_sr_Hind3-672953 |
inoculation of pCEP4
Investigators: Tom S.
Time: 2012-09-04 6 pm
Materials:
- LB medium
- Amp 25 mg/ml stock solution
- plate with cultures: pCEP4 (from 2012-09-03)
Method:
Inoculation of:
1 culture of pCEP4 plate in 5 ml LB medium + 5 µL amp.
(--> 1 culture)
Further tasks:
- Miniprep
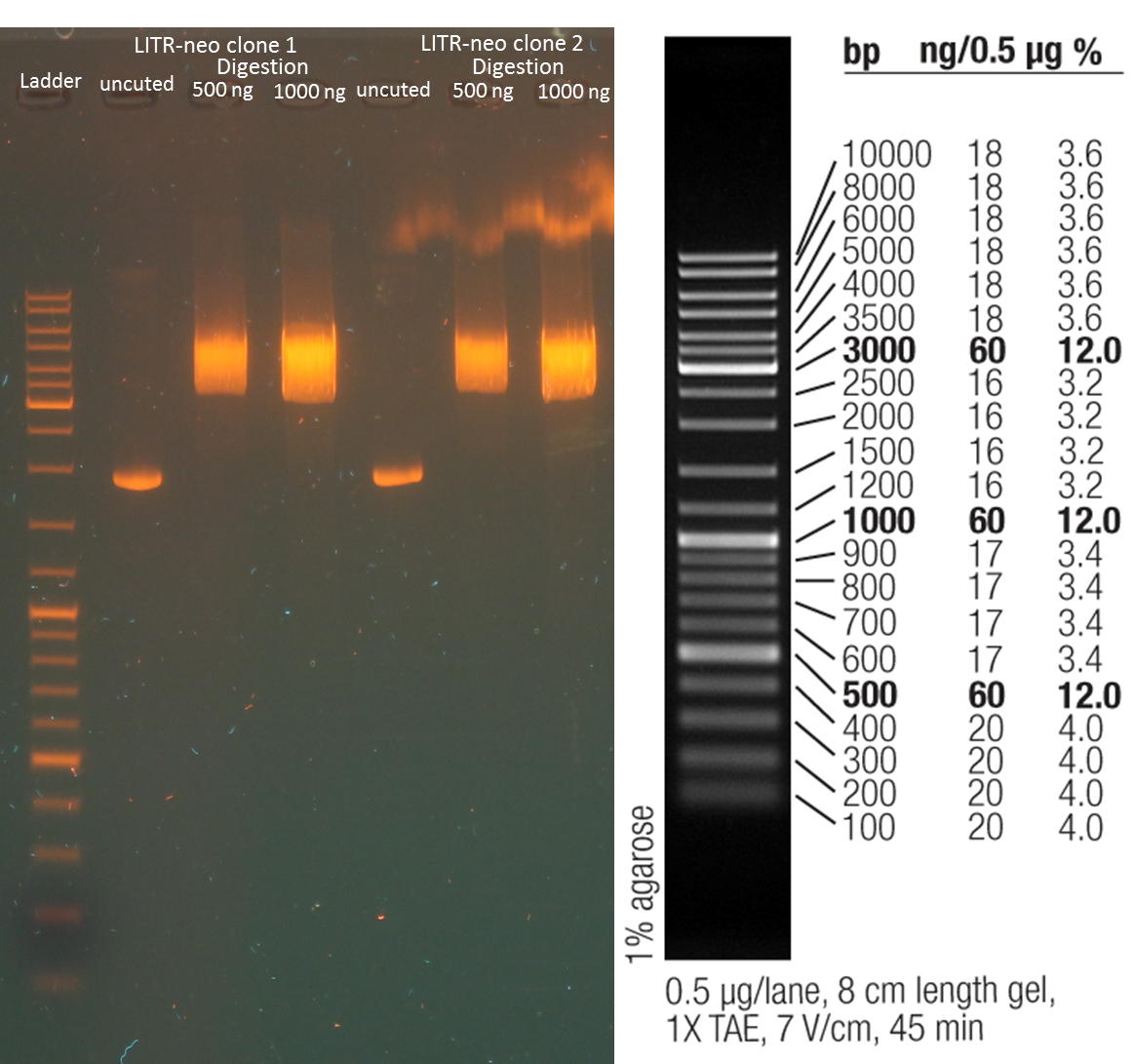
repetition of digestion of pAK100& scFV pKMEF425bla (SfiI&AscI)
Investigators: Chris
Time: 2012-09-04 9 pm
Method:
sample preparation:
5µL (2000 ng) pAK100, 20 µL H2O, 3 µL Neb3, 2µL AscI
25 µL ScFV, 3 µL Neb3, 2µL AscI
incubation over night at 37°C
Further tasks:
- addition of BSA, SfiI and incubation at 50 °C
Overnight culture of ER2738 cells
Investigators: Basia
Time: 2012-09-04 7 pm
Materials:
LB medium, tetracycline stock solution, glycerol stock of ER2738 cells
Method: inoculation in 20 ml LB medium + 20µl tetracycline stock, shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
preparation of competent cells and helper phage culturing
2012-09-05
Preparation of helper phage
Investigators: Basia/Chris
Time: 2012-09-05 10am
Materials:
LB medium, tetracycline stock solution, overnight culture of ER2738 cells, helper phage stock solution, Kanamycin, PEG-NaCl solution, TBS buffer
Method:
amplification and clean up of helper phage according to the manual
Further tasks:
continuation on the next day
Preparation of competent cells ER2738
Investigators: Basia/Chris
Time: 2012-09-05 10am
Materials:
LB medium, tetracycline stock solution, overnight culture of ER2738 cells, CaCl2, Glycerol
Method:
preparation of competent cells according to the manual
Results:
competent cells ready to use
Further tasks:
Transformation
Transformation of pBad with wtAID
Investigators: Basia
Time: 2012-09-05 6pm
Materials:
- Bunsen burner, Agar plates with ampicillin
- icebox
- competent ER2738 cells
- pBad plasmid with wtAID
Method:
Transformation via manual, 5 µl of plasmid was used
Plate incubation start: 7:30 pm
Results:
ready mutants to pick clones
Further tasks:
picking clones
Documentation of growing CHO cells under the microscope (24h incubation)
Investigators: Rico, Tom S.
Time: 2012-09-05
Materials:
- six well plates with growing CHO cells (24h), which were transfected with EGFR-construct alone or in combination with WT AID or AID without NES, with NLS+Kozak sequence or AID without NES, with NLS+Kozak sequence+eGFP
- Fluorescence microscope
Method:
take a photo of each well with a filter for GFP, YFP and brightfield
Isolation of mutation rate samples (EGFR-construct (EGFR-C) alone, with AID without NES, with NLS+Kozak sequence, with AID without NES, with NLS+Kozak sequence+eGFP and with wildtype AID from transfected CHO-Cells (24h incubation))
Investigators:
Rico, Tom S.
Aim:
- Isolation of mutation rate samples (EGFR-construct alone, with AID without NES, with NLS+Kozak sequence, with AID without NES, with NLS+Kozak sequence+eGFP and with wild type AID from transfected CHO cells) to separate the plasmids for sequencing
Method:
- Miniprep of cells
- Elution with 20 µL
Results:
CHO-transfected with EGFR-C = 20,7 ng/µL
CHO-transfected with EGFR-C and WT AID = 18,1 ng/µL
CHO-transfected with EGFR-C and AID without NES, with NLS+Kozak sequence = 55,7 ng/µL
CHO-transfected with EGFR-C and AID without NES, with NLS+Kozak sequence+eGFP = 40,0 ng/µL
Further tasks:
transformation of the plasmids
Transformation of plasmids from cells which grew for 24h (Mutation rate)
Investigators: Tom
Time: 2012-09-05
Materials:
- Bunsen burner, Agar plates with chloramphenicol
- icebox
- competent E. coli cells (XL 1 Blue)
- samples of 24h incubation
Method:
Transformation via manual, 1:2 dilutions of prepped samples were used
Results:
ready for growing mutants
Further tasks:
picking clones
2012-09-06
Preparation of helper phage - continuation
Investigators: Basia
Time: 2012-09-06
Materials:
LB medium, PEG-NaCl solution, TBS buffer
Method:
continuation of clean up of helper phage according to the manual
Overnight culture of ER2738 cells with AID in pBAD
Investigators: Basia
Time: 2012-09-06 5:30 pm
Materials:
LB medium, ampicillin stock solution, plate with transformed colonies from 5.9.2012
Method: inoculation in 20 ml LB medium + 20µl ampicillin stock, shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
preparation of competent ER2738 cells with AID in pBAD
Documentation of growing CHO-Cells under the microscope (48h incubation)
Investigators: Rico, Tom S.
Time: 2012-09-06
Materials:
- six well plates with growing CHO cells (24h) which were transfected with EGFR-construct alone or in combination with WT AID or AID without NES, with NLS+Kozak sequence or AID without NES, with NLS+Kozak sequence+eGFP
- Fluorescence microscope
Method:
take a photo of each well with a filter for GFP, YFP and brightfield
Isolation of mutation rate samples (EGFR-construct (EGFR-C) alone, with AID without NES, with NLS+Kozak sequence, with AID without NES, with NLS+Kozak sequence+eGFP and with wild type AID from transfected CHO cells (48h incubation))
Investigators:
Rico, Tom S.
Aim:
- Isolation of mutation rate samples (EGFR-construct alone, with AID without NES, with NLS+Kozak sequence, with AID without NES, with NLS+Kozak sequence+eGFP and with wild type AID from transfected CHO cells) to separate the plasmids for sequencing
Method:
- Miniprep of cells
- Elution with 20 µL
Results:
CHO-transfected with EGFR-C = 45,9 ng/µL
CHO-transfected with EGFR-C and WT AID = 41,7 ng/µL
CHO-transfected with EGFR-C and AID without NES, with NLS+Kozak sequence = 24,8 ng/µL
CHO-transfected with EGFR-C and AID without NES, with NLS+Kozak sequence+eGFP = 18,6 ng/µL
Further tasks:
transform plasmids
Inoculation of plasmid samples of the 24h retransformation plates
Investigators: Tom S.
Time: 2012-09-06 4pm
Materials:
- LB medium
- Amp 25 mg/ ml stock solution
- plate with cultures: EGFR-C
EGFR-C-WT AID
EGFR-C-AID without NES, with NLS+Kozak sequence
EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP
(all from 2012.09.06)
Method:
Inoculation of:
5 cultures per plate in 5 ml LB medium + 5µL amp.
(--> 20 cultures)
Further tasks:
- Miniprep
Checking the sequencing data
Investigators: Tom S.
Aim: choose the right constructs
Results:
CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH in pSB1C3 2-1-2 -> is good
2012-09-07
Preparation of competent cells ER2738+AID in pBAD
Investigators: Basia
Time: 2012-09-07 10am
Materials:
LB medium, ampicillin stock solution, overnight culture of ER2738 cells with AID in pBAD, CaCl2, Glycerol
Method:
preparation of competent cells according to the manual
Results:
competent cells ready to use
Further tasks:
Transformation
Transformation of pAK100 with scFV into ER2738 with AID in pBAD
Investigators: Basia
Time: 2012-09-07 7pm
Materials:
- Bunsen burner, Agar plates with ampicillin
- icebox
- competent ER2738 cells
- pAK100 plasmid with scFV L1
Method:
Transformation via manual, 4 µl of plasmid was used
Plate incubation start: 7:00 pm
Results:
ready mutants to pick clones
Further tasks:
picking clones
Miniprep of overnight cultures of 24h cultures for mutation rates
Investigators:
Rico, Tom S.
Aim:
Miniprep of overnight cultures of 24h cultures for mutation rates
Method:
Miniprep Thermo Scientific according to the manual
Results:
Concentrations
| Sample | Concentration [ng/µL] |
| EGFR-C 1 | 378,3 |
| EGFR-C 2 | 318,0 |
| EGFR-C 3 | 385,6 |
| EGFR-C 4 | 303,9 |
| EGFR-C 5 | 368,6 |
| EGFR-C-WT AID 1 | 274,9 |
| EGFR-C-WT AID 3 | 393,8 |
| EGFR-C-WT AID 4 | 313,5 |
| EGFR-C-WT AID 5 | 299,6 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 1 | 232,7 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 2 | 376,5 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 3 | 399,2 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 4 | 394,2 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 5 | 243,0 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 1 | 408,4 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 2 | 391,5 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 3 | 440,2 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 4 | 364,5 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 5 | 468,3 |
Further Tasks:
send to sequencing
send the DNA to sequencing for mutation rate (24h)
Investigators:
Tom S.
| sample | GATC number | Seq. Primer |
| EGFR-C 1 | II3641 | pcDNA 3.1-FP |
| EGFR-C 2 | II3642 | pcDNA 3.1-FP |
| EGFR-C 3 | II3643 | pcDNA 3.1-FP |
| EGFR-C 4 | II3644 | pcDNA 3.1-FP |
| EGFR-C 5 | II3645 | pcDNA 3.1-FP |
| EGFR-C-WT AID 1 | II3646 | pcDNA 3.1-FP |
| EGFR-C-WT AID 3 | II3647 | pcDNA 3.1-FP |
| EGFR-C-WT AID 4 | II3648 | pcDNA 3.1-FP |
| EGFR-C-WT AID 5 | II3649 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence 1 | II3650 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence 2 | II3651 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence 3 | II3652 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence 4 | II3653 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence 5 | II3654 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 1 | II3655 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 2 | II3656 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 3 | II3657 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 4 | II3658 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 5 | II3659 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP pool | II3660 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence pool | II3661 | pcDNA 3.1-FP |
Documentation of growing CHO cells under the microscope (72h incubation)
Investigators: Rico, Tom S.
Time: 2012-09-07
Materials:
- six well plates with growing CHO cells (72h) which were transfected with EGFR-construct alone or in combination with WT AID or AID without NES, with NLS+Kozak sequence or AID without NES, with NLS+Kozak sequence+eGFP
- Fluorescence microscope
Method:
take a photo of each well with a filter for GFP, YFP and brightfield
Isolation of mutation rate samples (EGFR-construct (EGFR-C) alone, with AID without NES, with NLS+Kozak sequence, with AID without NES, with NLS+Kozak sequence+eGFP and with wild type AID from transfected CHO cells (72h incubation))
Investigators:
Rico
Aim:
- Isolation of mutation rate samples (EGFR-construct alone, with AID without NES, with NLS+Kozak sequence, with AID without NES, with NLS+Kozak sequence+eGFP and with wild type AID from transfected CHO cells) to separate the plasmids for sequencing
Method:
- Miniprep of cells
- Elution with 50 µL
Results:
CHO-transfected with EGFR-C = 16,3 ng/µL
CHO-transfected with EGFR-C and WT AID = 15,3 ng/µL
CHO-transfected with EGFR-C and AID without NES, with NLS+Kozak sequence = 23,9 ng/µL
CHO-transfected with EGFR-C and AID without NES, with NLS+Kozak sequence+eGFP = 21,5 ng/µL
Further tasks:
transform plasmids
Transformation of samples from cells which grew for 48h and 72h (Mutation rate)
Investigators:Rico, Tom S.
Time: 2012-09-07
Materials:
- Bunsen burner, Agar plates with chloramphenicol
- icebox
- competent E. coli cells (XL 1 Blue)
- samples of 48h and 72h incubation
Method:
Transformation via manual, 1:5 dilutions of miniprep samples were used
Results:
ready for growing mutants
Further tasks:
picking clones
2012-09-08
Inoculation of plasmid samples of the 48h retransformation plates
Investigators: Basia
Time: 2012-09-08 6pm
Materials:
- LB medium
- Amp 25 mg/ ml stock solution
- plate with cultures: EGFR-C
EGFR-C-WT AID
EGFR-C-AID without NES, with NLS+Kozak sequence
EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP
(all from 2012.09.07)
Method:
Inoculation of:
5 cultures per plate in 5 ml LB medium + 5µL amp.
(--> 20 cultures)
Further tasks:
- Miniprep
Inoculation of cells ER2738 with AID in pBAD and scFV in pAK100
Investigators: Basia
Time: 2012-09-08 11am
Materials:
LB medium, ampicillin stock solution, chloramphenicol stock solution, arabinose 10% stock solution, plates with ER2738 cells with AID in pBAD and scFV in pAK100,
Method:
picking clones from a plate with ER2738 cells with AID in pBAD and scFV in pAK100 into 200ml of LB medium with antibiotics (ampicillin and chloramphenicol) and 0,1% arabinose or 0,01% arabinose (2 flasks, 200ml LB in each)
Results:
Slow growth of the cells
Further tasks:
Inoculation of overnight culture without arabinose
Inoculation of cells ER2738 with AID in pBAD and scFV in pAK100
Investigators: Basia
Time: 2012-09-08 5pm
Materials:
LB medium, ampicillin stock solution, chloramphenicol stock solution, plates with ER2738 cells with AID in pBAD and scFV in pAK100,
Method:
picking clones from a plate with ER2738 cells with AID in pBAD and scFV in pAK100 into 5ml of LB medium with antibiotics (ampicillin and chloramphenicol)
Results:
culture grew
Further tasks:
preparation of phages
PCR of AID with Thiophosphate primers
Investigators: Rico
Materials:
Mastermix
| reagent | volume [µL] |
| HF Phusion buffer 5x | 10 |
| dNTPs | 1 |
| Primer (Forward) | 1,25 |
| Primer (Reverse) | 1,25 |
| DNA (BBa_K103001 AID in pSB1A2 10 ng/µl) | 1,0 |
| Phusion Polymerase | 0,5 |
| water | 35,0 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 5 | 17 |
| annealing | 66 | 20 | 17 |
| elongation | 72 | 18 | 17 |
| denaturation | 98 | 5 | 17 |
| annealing+elongation | 72 | 18 | 17 |
| final elongation | 72 | 600 | 1 |
| cooling | 4 | ∞ | 1 |
Further Tasks:
PLICing variants
Digestion of wt AID
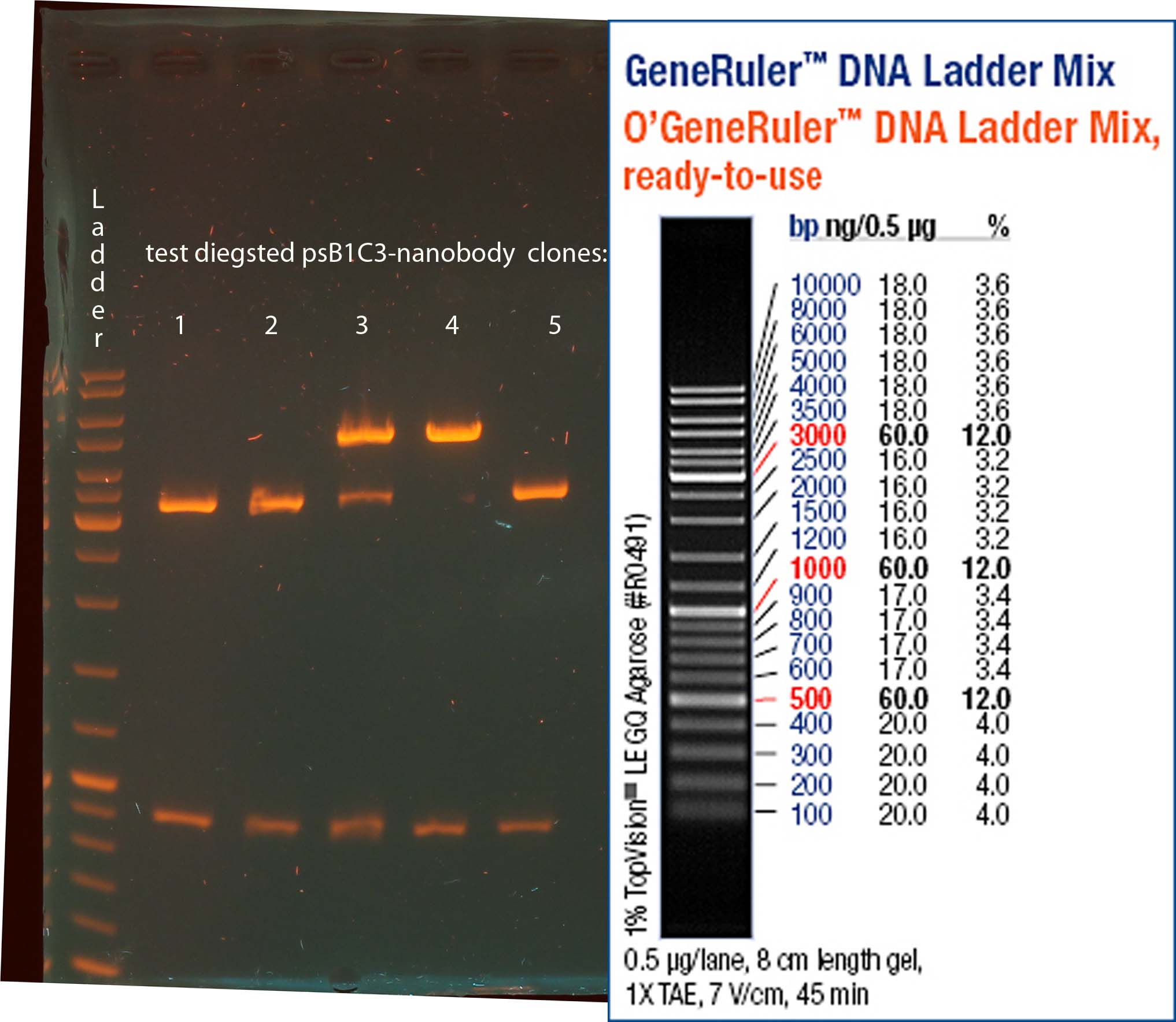
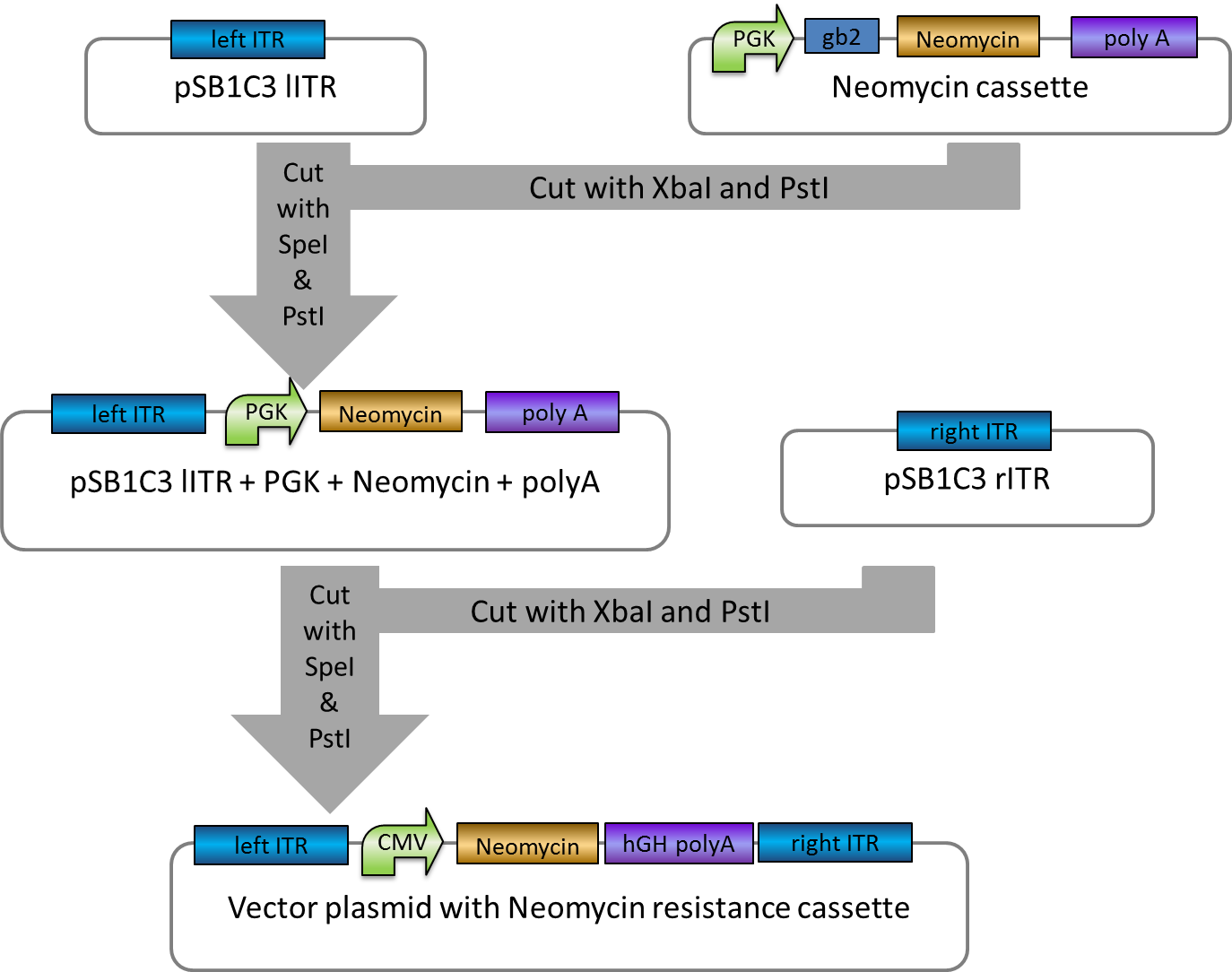
Investigators: Rico
Aim: get the backbone for the Potsdam standard cloning vector
Materials:
- digestion with 1 µL XbaI and 1 µL PstI
Further Tasks:
- electrophoretic separation
- ligation of the new standard cloning vector
2012-09-09
Preparation of the phages for phage display
Investigators: Basia
Time: 2012-09-09 10am-11pm
Materials:
LB medium, tetracycline stock solution, chloramphenicol stock solution, overnight culture of ER2738 cells, helper phage, Kanamycin, PEG-NaCl solution, TBS buffer
Method:
1. 2 Erlenmeyer flasks 100ml LB in each + 100µl of ampicillin stock solution + 100µl of chloramphenicol stock solution
2. one with no arabinose, the other one with 0,01% arabinose (when OD600 0,3-0,5)
3. addition of 30µl of helper phages (cleaned up on 6.9.2012) - when OD600 0,3-0,5
4. further amplification and clean up of phage according to the manual
Further tasks:
continuation on the next day
Miniprep of overnight cultures of 48h cultures for mutation rates
Investigators:
Basia
Aim:
Miniprep of overnight cultures of 48h cultures for mutation rates
Method:
Miniprep Thermo Scientific according to the manual
Results:
Concentrations
| Sample | Concentration [ng/µL] |
| EGFR-C 1 | 201.6 |
| EGFR-C 2 | 110.1 |
| EGFR-C 3 | 106.1 |
| EGFR-C 4 | 180.0 |
| EGFR-C 5 | 128.1 |
| EGFR-C-WT AID 1 | 121,1 |
| EGFR-C-WT AID 2 | 118,4 |
| EGFR-C-WT AID 3 | 136,1 |
| EGFR-C-WT AID 4 | 110.8 |
| EGFR-C-WT AID 5 | 122,1 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 1 | 157,1 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 2 | 125,3 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 3 | 133,9 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 4 | 176,5 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 5 | 142,3 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 1 | 112,2 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 2 | 125,1 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 3 | 107,8 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 4 | 123,6 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 5 | 114,2 |
Further Tasks:
send to sequencing
Inoculation of plasmid samples of the 72h retransformation plates
Investigators: Basia
Time: 2012-09-09 6pm
Materials:
- LB medium
- Amp 25 mg/ ml stock solution
- plate with cultures: EGFR-C
EGFR-C-WT AID
EGFR-C-AID without NES, with NLS+Kozak sequence
EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP
(all from 2012.09.07)
Method:
Inoculation of:
5 cultures per plate in 5 ml LB medium + 5µL amp.
(--> 20 cultures)
Further tasks:
- Miniprep
Inoculation of cells ER2738 with AID in pBAD and cells ER2738 without AID in pBAD
Investigators: Basia
Time: 2012-09-08 8:30pm
Materials:
LB medium, ampicillin stock solution, tetracyclin stock solution, competent ER2738 cells with AID in pBAD and competent ER2738 cells without AID in pBAD
Method:
100µl of competent ER2738 cells with or without AID in pBAD and scFV in pAK100 into 5ml of LB medium with antibiotics (ampicillin or tetracyclin)
Results:
cultures grew
Further tasks:
infection with phages and selection for mutated clones
2012-09-10
Preparation of phage - continuation
Investigators: Basia/Chris
Time: 2012-09-10
Materials:
LB medium, PEG-NaCl solution, TBS buffer
Method:
continuation of clean up of phages for phage display
Infection with phages of the cells ER2738 with AID in pBAD and without AID in pBAD
Investigators: Basia /Chris
Time: 2012-09-10 2pm
Materials:
LB medium, tetracycline stock solution, chloramphenicol stock solution, ampicillin stock solution, overnight culture of ER2738 cells (once with once without AID in pBAD), phages cleaned up on 10.09.2012
Method:
1. 4 Erlenmeyer flasks 100ml LB in each + 100µl of ampicillin stock solution (into the flasks with AID in pBAD) - x2 or + 100µl of tetracycline stock solution (into flasks without AID) - x2
2. two with no arabinose (into the flasks without AID in pBAD), the other two with 0,01% arabinose (when OD600 0,3-0,5, into flasks with AID)
3. addition of 280µl of phages (cleaned up on 10.9.2012) - when OD600 0,3-0,5
4. addition of chloramphenicol stock solution - 1h after infection with phages
5. further amplification of infected cells in 32°C
Further tasks:
Plating of the colonies onto the LB plates with appropriate antibiotics
Plating of the colonies onto the LB plates with appropriate antibiotics
Investigators: Chris
Time: 2012-09-10 9pm
Materials:
Plates with LB medium with tetracycline and chloramphenicol and plates with LB medium with ampicillin and chloramphenicol, cultures infected with phages
Method:
850µl of the cultures infected with phages (see 10.09.2012 - phage infection) were centrifuged and the supernatant was discarded. 20µl of the resuspended pellet was used for plating
each culture was plated on the plates with appropriate antibiotics (E. coli without AID -infected with Phages 1&2 on Tet and and Chloramphenicol, E. coli with AID -infected with Phages 1&2 on Chloramphenicol and amp)
Further tasks:
preparation of mutated vectors for sequences
Purification of transformed plasmids from the transfected CHO cells
Aim: purification of the transformed single chain construct with YFP from the CHO cells 3 days after transfection
Investigators: Maria, Rico
Method: purification kit from Thermo Scientific
Results:
Concentrations
| Sample | Concentration [ng/µL] |
| EGFR-C 1 | 367.3 |
| EGFR-C 2 | 342.6 |
| EGFR-C 3 | 276.0 |
| EGFR-C 4 | 285.1 |
| EGFR-C 5 | 263.3 |
| EGFR-C-WT AID 1 | 288.3 |
| EGFR-C-WT AID 2 | 301.4 |
| EGFR-C-WT AID 3 | 292.7 |
| EGFR-C-WT AID 4 | 317.9 |
| EGFR-C-WT AID 5 | 293.4 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 1 | 373.1 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 2 | 356.8 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 3 | 399.4 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 4 | 357.1 |
| EGFR-C-AID without NES, with NLS+Kozak sequence 5 | 325.4 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 1 | 315.4 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 2 | 305.6 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 3 | 359.5 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 4 | 330.2 |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 5 | 374.2 |
2012-09-11
picking clones and overnight culture of the colonies grown on the plates from 10.9.2012 and XL1 BLue with CFP and YFP
Investigators: Basia/Chris
Time: 2012-09-03 7 pm
Materials:
LB medium, ampicillin stock solution, chloramphenicol stock solution, tetracycline stock solution, plates with colonies from 10.9.2012
Method: inoculation in 5 ml LB medium + 5µl ampicillin chloramphenicol & tetracycline:
the only colony of ER2738 with AID infected with Phage 1 inoculated
inoculation in 5 ml LB medium + 5µl chloramphenicol & tetracycline:
ER2738 without AID infected with Phage 1 and 2-> 5 single colonies, 1 mixed culture (500 µl LB were spread on the plate and 10 µL used for inoculation)of each
inoculation in 50 ml LB medium + 50 µl ampicillin
XL1-Blue with CFP and Xl1-Blue with YFP
shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
Miniprep, inoculation of overnight cultures (CFP, YFP, Mixed cultures - P1-AID, P1+AID, P2-AID)
Transfection of CHO cells in 6 well plate and ibidi dishes
Investigator: Rico
Aim: transfection of CHO cells with EGFR-C/modified AID, EGFR-C/ wt AID and EGFR-C alone in 6 well dishes and modified AID alone in ibidi dishes
Further Tasks: documentation of fluorescence with fluorescence microscope
PCR of RFP with restriction sites for RFC 10
Investigator: Sascha, Rico
Aim: amplification of RFP to add the restriction sites for RFC 10
Materials:
Mastermix
| reagent | volume [µL] |
| HF Phusion buffer 5x | 10 |
| dNTPs | 1 |
| Primer (Forward) | 1,25 |
| Primer (Reverse) | 1,25 |
| DNA (BBa_K103001 AID in pSB1A2 10 ng/µl) | 1,0 |
| Phusion Polymerase | 0,5 |
| water | 35,0 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 5 | 17 |
| annealing | 70 | 20 | 17 |
| elongation | 72 | 18 | 17 |
| denaturation | 98 | 5 | 17 |
| annealing+elongation | 72 | 18 | 17 |
| final elongation | 72 | 600 | 1 |
| cooling | 4 | ∞ | 1 |
Furhter Tasks: ligation of RFP and digested pSB1C3
2012-09-12
Miniprep of overnight cultures
Investigators:
Chris/Basia
Aim:
Miniprep of overnight cultures
Method:
Miniprep Thermo Scientific according to the manual
Results:
Concentrations
| Sample | Concentration [ng/µL] |
| CFP (AmpR) | 184 |
| Phage 1 C1 | 740 |
| Phage 1 C2 | 843 |
| Phage 1 C3 | 740 |
| Phage 1 C4 | 620 |
| Phage 1 C5 | 685 |
| Phage 2 C1 | 842 |
| Phage 2 C2 | 689 |
| Phage 2 C3 | 622 |
| Phage 2 C4 | 683 |
| Phage 2 C5 | 777.6 |
Phage 1 was produced under expression of AID (mutated, but there was no AID while reinfection of E.coli)
Phage 2 is the control(Phage production and Infection without AID)
C1-C5 where picked colonies
send DNA from colonies after first round of phage display to sequencing
Investigators:
Basia/Chris
| sample | GATC number | Seq. Primer |
| Phage1(mut) C1 | II3662 | GATC_Std_RPC |
| Phage1(mut) C2 | II3663 | GATC_Std_RPC |
| Phage1(mut) C3 | II3664 | GATC_Std_RPC |
| Phage1(mut) C4 | II3665 | GATC_Std_RPC |
| Phage1(mut) C5 | II3666 | GATC_Std_RPC |
| Phage2(not mut) C1 | II3667 | GATC_Std_RPC |
| Phage2(not mut) C2 | II3668 | GATC_Std_RPC |
| Phage2(not mut) C3 | II3669 | GATC_Std_RPC |
| Phage2(not mut) C4 | II3670 | GATC_Std_RPC |
| Phage2(not mut) C5 | II3671 | GATC_Std_RPC |
Inoculation of main cultures: CFP (ampR), mixed colonies of mutated and non mutated infected colonies (in E.coli without AID ->Tet, CM-resistance)
Investigators:Basia/Chris
Aim:
Purfication of CFP protein, determination of mutation rates via phage display
Materials:
LB medium, tetracycline stock solution, chloramphenicol stock solution, ampicillin stock solution, overnight culture of CFP producing cells, overnight culture of E. coli infected with phages
Method:
Preparation of CFP main culture:
2x2ml of overnight culture was added to 2x 500ml LB medium with 500µl of ampicillin each
Preparation of main cultures for phage display:
1x 1ml of overnight culture of non-mutated cells were added to 100ml of LB medium with 100µl of tetracycline
1x 1ml of overnight culture of 1 x mutated cells were added to 100ml of LB medium with 100µl of tetracycline and 100µl of ampicillin
Further tasks:
Induction of CFP; new cycle of phage production
Induction of CFP
Investigators:Basia/Chris
Aim:
Purfication of CFP protein
Materials:
Main culture XL1 Blue with CFP, IPTG stock solution
Method:
250µl of 1M IPTG stock solution was added to each of 500ml of main cultures when OD600 reached 0,3-0,5
Further tasks:
protein purification
Preparation of the phages for phage display
Investigators: Basia/Chris
Time: 2012-09-12
Materials:
main culture of ER2738 with pBAD, pAK100 , helper phage, Kanamycin, PEG-NaCl solution, TBS buffer
Method:
1. addition of arabinose to the culture with pBAD with AID: 0,01% arabinose (when OD600 0,3-0,5)
3. addition of 30µl of helper phages (cleaned up on 6.9.2012) - when OD600 0,3-0,5
4. further amplification and clean up of phage according to the manual
Further tasks:
continuation on the next day
Inoculation of cells ER2738 with AID in pBAD and cells ER2738 without AID in pBAD
Investigators: Basia/Chris
Time: 2012-09-12 8:30pm
Materials:
LB medium, ampicillin stock solution, tetracyclin stock solution, competent ER2738 cells with AID in pBAD and competent ER2738 cells without AID in pBAD
Method:
100µl of competent ER2738 cells with or without AID in pBAD into 5ml of LB medium with antibiotics (ampicillin or tetracyclin)
Results:
cultures grew
Further tasks:
infection with phages and selection for mutated clones
send the DNA to sequencing
Investigators:
Tom S.
| sample | GATC number | Seq. Primer |
| mod. AID+eGFP 4 | II3676 | pSB1C3 Forward |
| mod. AID+eGFP 4 | II3677 | pSB1C3 Reverse |
Ligation of RFP in pSB1C3 and transformation
Investigators: Rico
Aim: Ligation of RFP in pSB1C3
Method: 1 µL RFP, 1 µL digested pSB1C3, 1 µL T4-Ligase, 1 µL T4-Ligation buffer, 6 µL water, transformation in Xl 1 E.coli (E.coli were spread without IPTG)
Further Tasks:
O/N culture of clones
2012-09-13
Preparation of phage - continuation
Investigators: Basia/Chris
Time: 2012-09-13
Materials:
LB medium, PEG-NaCl solution, TBS buffer
Method:
continuation of clean up phages for phage display according to the manual
Isolation of CFP from XL1-Blue
Investigators: Basia/Chris
Materials:
Method:
-resuspend pellet (in 20 mL 10mM Hepes 500mM NaCl Buffer pH 7.4)
-lysis by sonication: 3x2min, 6mm tip, 50% amplitude
-centrifugation´for 30 min, 19000 rpm 4°C
-sonication of supernatant for 1 min
-filtering with 0.45 mm filter
-loading extract on IMAC-column(Ni-NTA) (equilibrated with 30 column volumes of 10 mM HEPES buffer-pH 7.4; 500mM NaCL)
-loading protein extract with flow rate of 0.5 mL/min
-wash column with 30mM imidazole
-elution of protein with 50 mM imidazole in 10 mM HEPES bufer pH 7.4 (this was a mistake-200mM imidazole is the correct elution buffer, but it worked anyways)
-storage of eluted protein on ice in the fridge
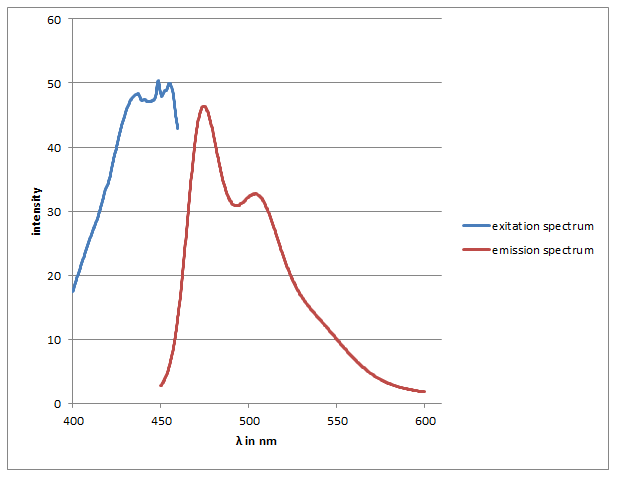
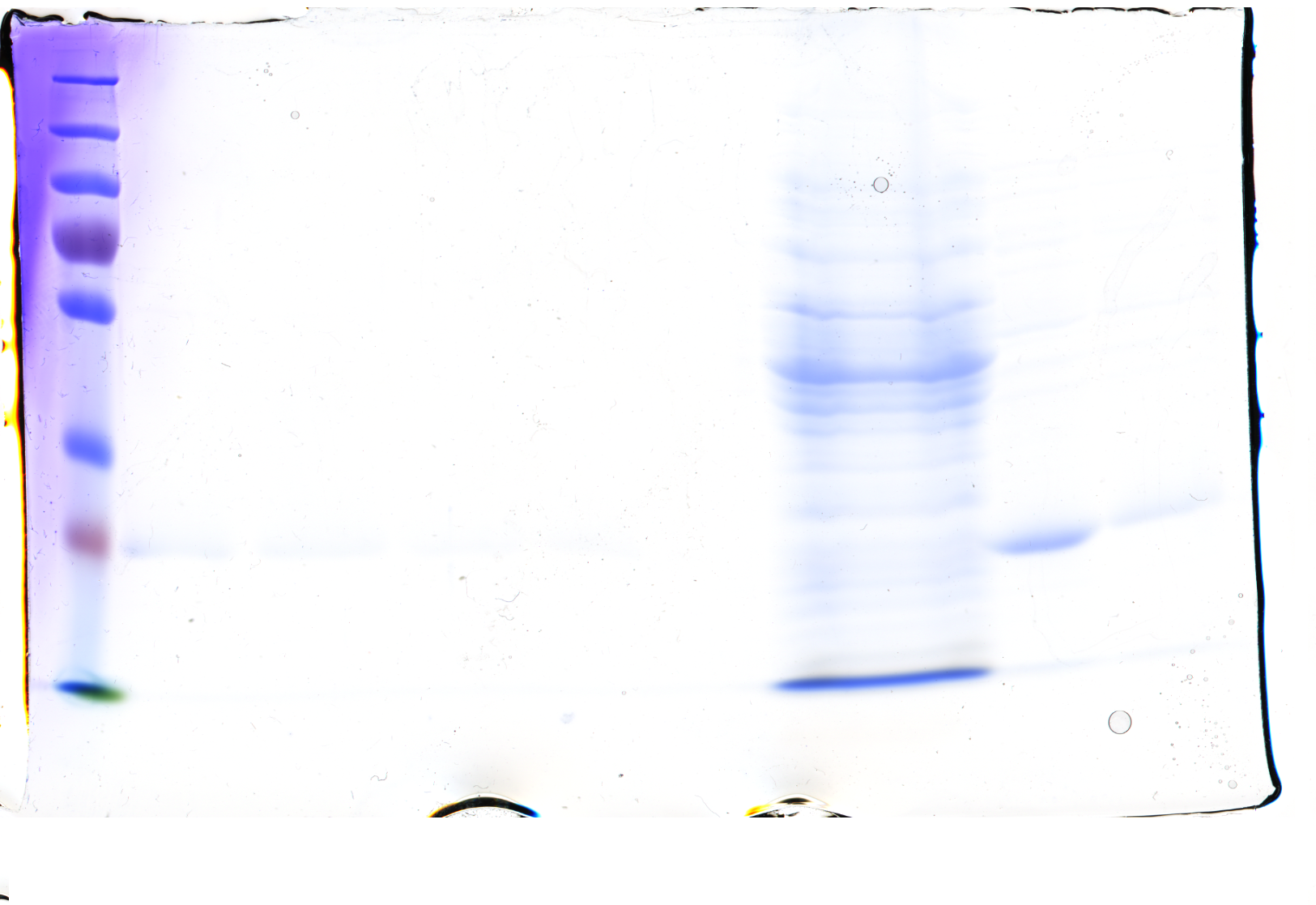
checking the SDS-Gel & spectrum of CFP
Investigators: Basia/Chris
Results:
-spectra look like CFP spectra (max. absorbtion at 436 nm) max emission 474 & nm
-in the strong diluted elution fraction the visible lane is as large as CFP (~28 kDA)
-this lane is also strong in the washed fraction
2012-09-14
send the DNA to sequencing for mutation rate (48h)
Investigators:
Tom S.
| sample | GATC number | Seq. Primer |
| EGFR-C 1 | AKM001W075 | pcDNA 3.1-FP |
| EGFR-C 2 | AKM001W076 | pcDNA 3.1-FP |
| EGFR-C 3 | AKM001W077 | pcDNA 3.1-FP |
| EGFR-C 4 | AKM001W078 | pcDNA 3.1-FP |
| EGFR-C 5 | AKM001W079 | pcDNA 3.1-FP |
| EGFR-C-WT AID 1 | AKM001W080 | pcDNA 3.1-FP |
| EGFR-C-WT AID 2 | AKM001W081 | pcDNA 3.1-FP |
| EGFR-C-WT AID 3 | AKM001W082 | pcDNA 3.1-FP |
| EGFR-C-WT AID 4 | AKM001W083 | pcDNA 3.1-FP |
| EGFR-C-WT AID 5 | AKM001W084 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence 1 | AKM001W085 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence 2 | AKM001W086 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence 3 | AKM001W087 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence 4 | AKM001W088 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence 5 | AKM001W089 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 1 | AKM001W090 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 2 | AKM001W091 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 3 | AKM001W092 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 4 | AKM001W093 | pcDNA 3.1-FP |
| EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP 5 | AKM001W094 | pcDNA 3.1-FP |
checking sequences after first round (1x mut) of phage display
Investigator: Chris
results: not a single mutation in one of the 5 "mutated sequences" or one of the 5 controls (without AID)
labeling GFP with Biotin (as a matter of fact GFP is fused with a helix-> CREBzipGFP)
Investigator: Basia/ Chris
Material: EZ-Link® Sulfo-NHS-LC-Biotin
Method:
-add 10 fold molar exess of biotin reagent to CREBzipGFP
-CREBzipGFP was thought to have an concentration of 1.3 mg/mL (MW=36.172 kDa), therefore 36 µL of 10 mM EZ-Link® Sulfo-NHS-LC-Biotin where added
-after biotinylation the concetration of CREBzipGFP-Biotin was shown to be 4.075 mg/mL -maybe to less Biotin reagent was used
-the concentration of remaining CREBzipGFP without Biotin was 0.3 mg/mL
Further tasks: testing the success of biotinylation with "ELISA"
inoculation of mixed plates ER2738-pAK100(not mutated) & ER2738-pAK100 with AID(2x mutated)
Investigator: Basia/ Chris
-addition of 1 mL LB medium on plates, mixing colonies with Drigalski spatulas
-addition of 20 µL of mixed cultures to 5 mL LB with Tet,CM (for ER2738+pAK100-not mutated) and to 5 mL LB with Tet,CM, Amp for ER2738+pAK100 with AID(2x mutated)
O/N cultures of RFP in pSB1C3 clones
Investigator: Rico
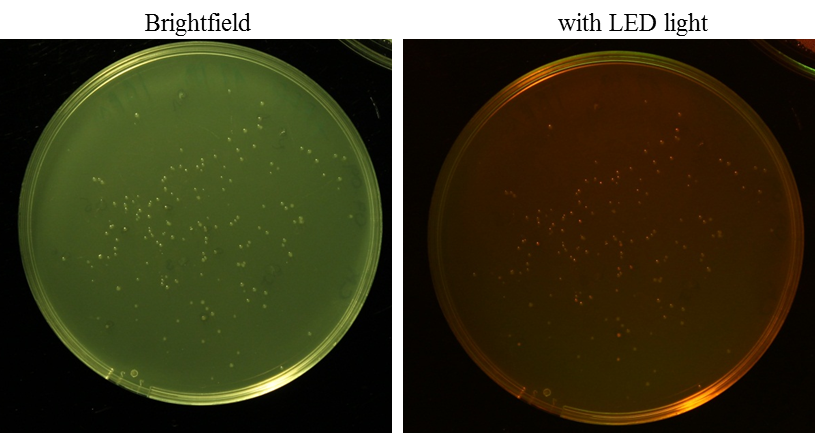
Aim: O/N culture of RFP in pSB1C3 clones, clones have to grow approximately 24 h at 37 °C and incubate O/N at 6-7 °C (after that you can see the red fluorescence under LED light)
Method: picking clones, 1:1000 dilution of IPTG stock solution (1 M)
Further Tasks: purification of plasmids
2012-09-15
Preparation of the phages for phage display
Investigators: Chris
Time: 2012-09-15
Materials:
over night cultures of ER2738 with pBAD and pAK(double mut) & ER2738 with pAK(not mut) , helper phage, Kanamycin, PEG-NaCl solution, TBS buffer
Method:
1. starting main culture of ER2738 with pBAD and pAK(double mut) & ER2738 with pAK(not mut)-addition of 2 mL over night culture to 100 mL LB(Tet,Cm for ER2738 with pAK /Tet,Cm, Amp for ER2738 with pBAD and pAK)
2. addition of arabinose to the culture with pBAD with AID: 0,02% arabinose (when OD600 0,3-0,5)
3. addition of 30µl of helper phages (cleaned up on 6.9.2012) - when OD600 0,3-0,5
4. further amplification and clean up of phage according to the manual
Further tasks:
continuation on the next day
overnight cultures of ER2738 and ER2738 with AID for phage display and AID-expression test
Investigator:Chris
Materials:
LB medium, ampicillin stock solution, tetracyclin stock solution, competent ER2738 cells with AID in pBAD and competent ER2738 cells without AID in pBAD
Method:
100µl of competent ER2738 cells with or without AID in pBAD into 5ml of LB medium with antibiotics (ampicillin & tetracyclin/ tetracyclin)
Results:
cultures grew
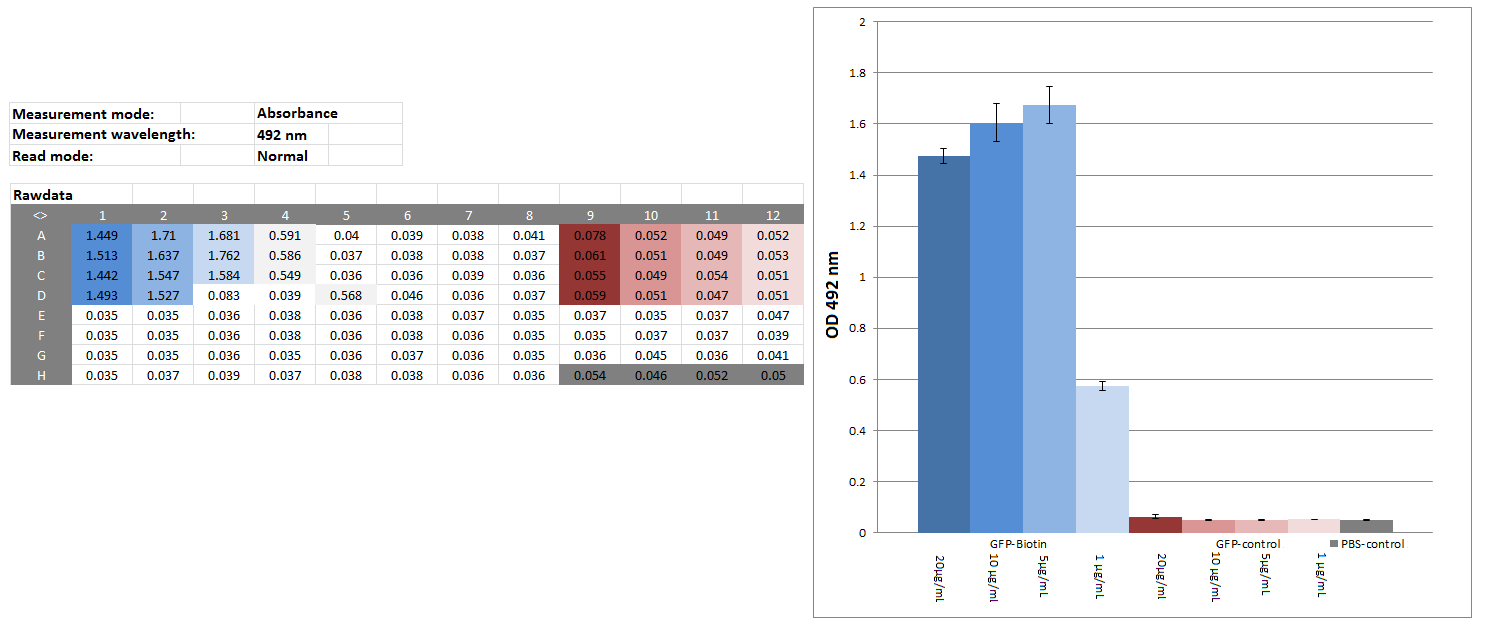
Elisa of biotinylated eGFP
Investigators: Chris
Materials:
CREBzipGFP-biotin (c=4.075 mg/mL), CREBzipGFP for control (c=0.3 mg/mL; ɛ280=22,015 M-1 cm-1; MW= 36,172 Da), block buffer (1% BSA in PBS), SAV-HPR-solution(SAV-HPR 1:5000 & 1&% BSA in PBS), substrate solution (20:78:1:1 5x --substrate Buffer: H20: Substrate->OPD: 1% H2O2), stop solution (1 M H2SO4,50mM Na2SO3 in water), platereader
Method:
CREBzipGFP-biotin (20 µg/mL,10 µg/mL,5 µg/mL, 1 µg/mL) CREBzipGFP-control (20 µg/mL,10 µg/mL,5 µg/mL, 1 µg/mL), PBS as blank: 50 µL/well
wait 2:30 h
washed 2 times w/tap water
block: 100 µL/well 1% BSA/PBS
washed 5 times w/tap water
50 µL/well SAV-HRP-solution, 1h
washed 10 times w/tap water
50 µL/well substrate solution - 5,5 min
50 µL/well stop solution,
measure OD 492
Results:
Further tasks:
couple CREBzipGFP-biotin with magnetic beads
Purification of RFP in pSB1C3 from O/N cultures
Investigator:Rico
Materials: Purification kit from Thermo Scientific
| sample | concentration in ng/µL |
| RFP 1 | 136.9 |
| RFP 2 | 113.8 |
| RFP 3 | 106.2 |
| RFP 4 | 99.8 |
| RFP 5 | 108.8 |
Digestion of purified new standard cloning vector with Apa I and Sph I
Investigator:Rico
Method:
7 µL RFP 1, 1 µL Apa I, 1 µL Sph I, 3 µL buffer, 18 µL water, incubation @ 37 °C for 2 h
Transformation of purified AID plasmids in E.coli for determining the mutation rate
Investigator:Rico
Materials: Transformation protocol for E.coli
2012-09-16
Induction calibration of AID in pBAD vector
Investigators: Basia
Time: 2012-09-16
Materials:
LB medium, protein gel equipment, ampicillin stock solution, tetracycline stock solution, arabinose stock solution
Method:
1. Inoculation of 200µl of overnight culture (ER2738+AID in pBAD) into 20ml of LB medium with tet and amp.
2. addition of arabinose 0,01%, when OD600 0,3-0,5
3. Samples taken at T0, T3 and T5 (at time zero, after 3h and after 5h)
4. The OD600 was measured before taking the samples in order to adjust the protein amount
5. Cell suspension was centrifuged for 3min 16100 xg and pellet was resuspended in 100µl of lyase sample buffer 4x.
Preparation of phage - continuation
Investigators: Basia
Time: 2012-09-16
Materials:
LB medium, PEG-NaCl solution, TBS buffer
Method:
continuation of clean up phages for phage display
Infection with phages of the cells ER2738 with AID in pBAD and without AID in pBAD
Investigators: Basia
Time: 2012-09-16 2pm
Materials:
LB medium, tetracycline stock solution, chloramphenicol stock solution, ampicillin stock solution, overnight culture of ER2738 cells (once with once without AID in pBAD), phages cleaned up on 16.09.2012, arabinose stock solution
Method:
1. 2 Erlenmeyer flasks 100ml LB in each + 100µl of ampicillin + 100µl tetracycline stock solution (into the flask with AID in pBAD) or + 100µl of tetracycline stock solution (into flask without AID) - x2
2. 1 with no arabinose (into the flask without AID in pBAD), the other one with 0,02% arabinose (when OD600 0,3-0,5, into flask with AID)
3. addition of 280µl of phages (cleaned up on 10.9.2012) - when OD600 0,3-0,5
4. addition of chloramphenicol stock solution - 1h after infection with phages
5. further amplification of infected cells in 32°C
Further tasks:
Plating of the colonies onto the LB plates with appropriate antibiotics
Plating of the colonies onto the LB plates with appropriate antibiotics
Investigators: Basia
Time: 2012-09-10 7pm
Materials:
Plate with LB medium with tetracycline and chloramphenicol and plates with LB medium with ampicillin, tetracycline, 0,01% arabinose and chloramphenicol, cultures infected with phages
Method:
850µl of the cultures infected with phages (see 16.09.2012 - phage infection) were centrifuged and the supernatant was discarded. 20µl of the resuspended pellet were used for plating
each culture was plated on the plates with appropriate antibiotics (E. coli without AID -infected with non-mutated phages on Tet and Cm, E. coli with AID -infected with mutated phages on Cm, tet, ara and amp)
Further tasks:
preparation of mutated vectors for sequences
Inoculation of plasmid samples of the 48h retransformation plates
Investigators: Basia
Time: 2012-09-08 6pm
Materials:
- LB medium
- Cm 35 mg/ ml stock solution
- plate with cultures:
EGFR-C-WT AID
EGFR-C-AID without NES, with NLS+Kozak sequence
EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP
(all from 2012.09.15)
Method:
Inoculation of:
5 cultures per plate in 5 ml LB medium + 5µL Cm.
(--> 20 cultures)
Further tasks:
- Miniprep
2012-09-17
Miniprep of overnight cultures of 48h cultures for (AID) mutation rates
Investigators:
Tom, Rico
Aim:
Miniprep of overnight cultures of 48h cultures for (AID) mutation rates
Method:
Miniprep Thermo Scientific according to the manual
Results:
Concentrations
| Sample | Concentration [ng/µL] |
| AID without NES, with NLS+Kozak sequence 1 | 371.2 |
| AID without NES, with NLS+Kozak sequence 2 | 368.4 |
| AID without NES, with NLS+Kozak sequence 3 | 340.8 |
| AID without NES, with NLS+Kozak sequence 4 | 391.3 |
| AID without NES, with NLS+Kozak sequence 5 | 373.9 |
| AID without NES, with NLS+Kozak sequence+eGFP 1 | 393.0 |
| AID without NES, with NLS+Kozak sequence+eGFP 2 | 413.1 |
| AID without NES, with NLS+Kozak sequence+eGFP 3 | 379.7 |
| AID without NES, with NLS+Kozak sequence+eGFP 4 | 376.5 |
| AID without NES, with NLS+Kozak sequence+eGFP 5 | 389.3 |
| WT (AID) 1 | 366.4 |
| WT (AID) 2 | 389.9 |
| WT (AID) 3 | 359.5 |
| WT (AID) 4 | 367.3 |
| WT (AID) 5 | 365.2 |
Further Tasks:
send to sequencing
send the DNA to sequencing
Investigators:
Tom S., Rico
| sample | GATC number | Seq. Primer |
| AID without NES, with NLS+Kozak sequence 1 | II3682 | CMV Forward |
| AID without NES, with NLS+Kozak sequence 2 | II3683 | CMV Forward |
| AID without NES, with NLS+Kozak sequence 3 | AI2677 | CMV Forward |
| AID without NES, with NLS+Kozak sequence 4 | AI2678 | CMV Forward |
| AID without NES, with NLS+Kozak sequence 5 | AI2679 | CMV Forward |
| AID without NES, with NLS+Kozak sequence+eGFP 1 | AI2680 | CMV Forward |
| AID without NES, with NLS+Kozak sequence+eGFP 4 | AI2681 | CMV Forward |
| AID without NES, with NLS+Kozak sequence+eGFP 3 | AI2682 | CMV Forward |
| AID without NES, with NLS+Kozak sequence+eGFP 4 | AI2683 | CMV Forward |
| AID without NES, with NLS+Kozak sequence+eGFP 5 | AI2684 | CMV Forward |
| WT (AID) 1 | AI2685 | CMV Forward |
| WT (AID) 2 | AI2686 | CMV Forward |
| WT (AID) 3 | AI2687 | CMV Forward |
| WT (AID) 4 | AI2688 | CMV Forward |
| WT (AID) 5 | AI2689 | CMV Forward |
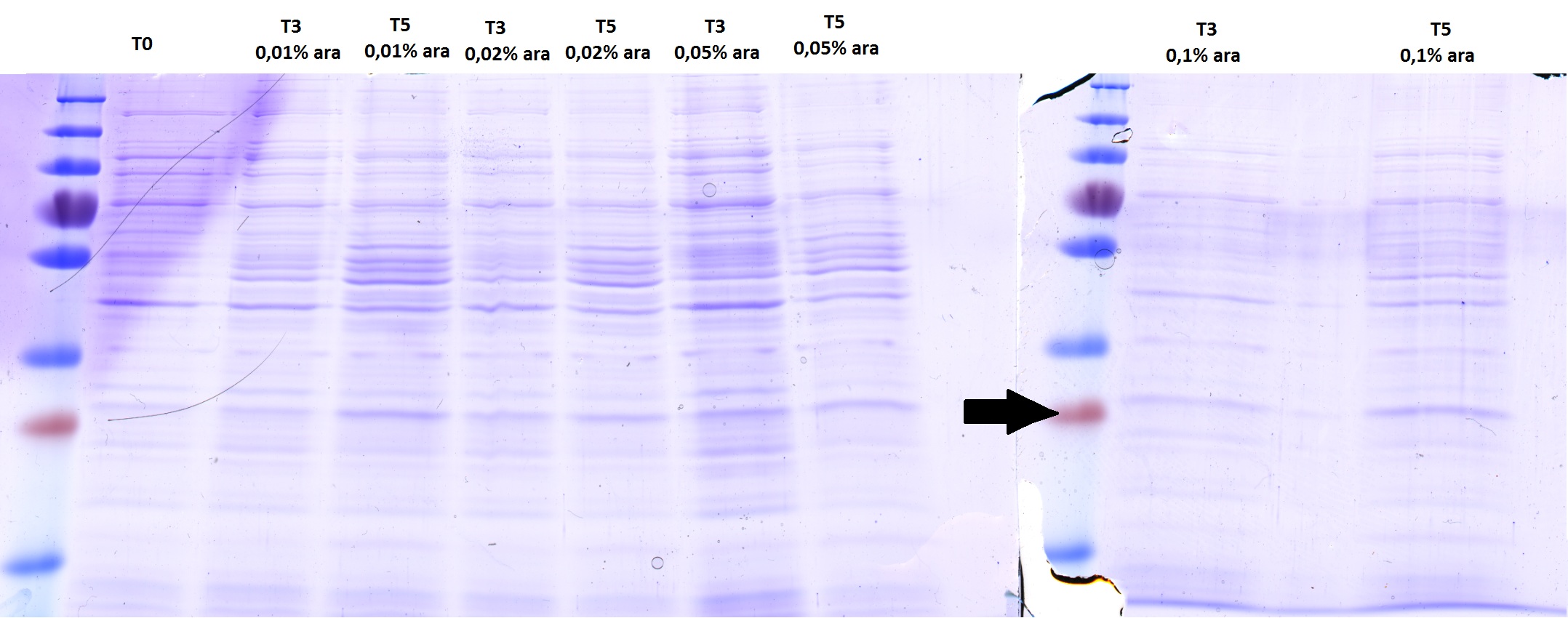
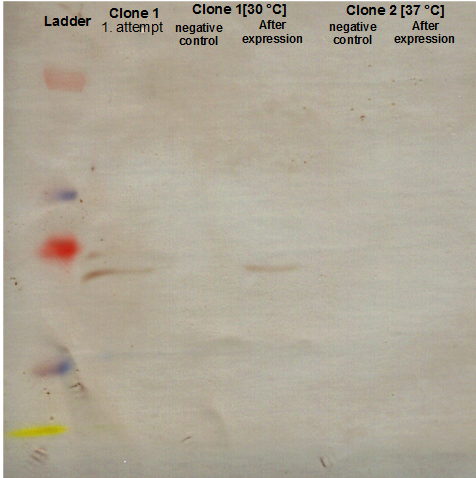
Checking AID induction test with SDS-PAGE gel
Investigators: Basia, Chris
There is no overexpressed lane that differs in the induced samples. No AID expression or too less arabinose???
picking clones and overnight culture of the colonies grown on the plates from 16.9.2012
Investigators: Basia
Time: 2012-09-17 7 pm
Materials:
LB medium, ampicillin stock solution, chloramphenicol stock solution, tetracycline stock solution, plates with colonies from 16.9.2012
Method: inoculation of 5 clones from mutated clones in 5 ml LB medium + 5µl ampicillin, chloramphenicol & tetracycline:
inoculation of 5 clones from the non mutated plate in 5 ml LB medium + 5µl chloramphenicol & tetracycline:
inoculation of XL1-Blue with AID in pBAD in 5 ml LB medium + 5 µl ampicillin + 5µl tetracycline - for a new induction test
shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
Miniprep, induction calibration
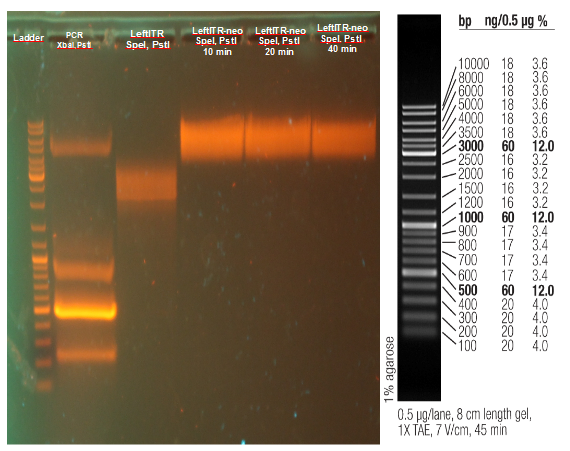
PLIcing of Thio-AID, Ligation with digested new RFC standard cloning vector 1
Investigators:
Rico
Aim:
Ligation of AID in new cloning vector 1
Method:
- 8 µL Thio-AID was treated with 1 µL Cleavage Buffer (0.5 Tris-HCl, pH 9.0), 0.4 µL Milli-Q water and 0.6 µL iodine stock solution (100 mM)
- incubate @ 70 °C for 5 min
- mixed with digested RFP standard cloning vector (purified, non purfied, with and without ligase) @ room temperature for 1 h
- transform into E.coli
2012-09-18
Induction calibration of AID in pBAD vector
Investigators: Basia/Chris
Time: 2012-09-18
Materials:
LB medium, protein gel equipment, ampicillin stock solution, tetracycline stock solution, arabinose stock solution
Method:
1. Inoculation of 200µl of overnight culture (ER2738+AID in pBAD) into 20ml of LB medium with tet and amp.
2. addition of arabinose 0,01%, when OD600 0,3-0,5
3. Samples taken at T0, T3 and T5 (at time zero, after 3h and after 5h)
4. The OD600 was measured before taking the samples in order to adjust the protein amount
5. Cell suspension was centrifuged for 3min 16100 xg and pellet was resuspended in 100µl of lyase sample buffer 4x.
Miniprep of overnight cultures
Investigators:
Chris/Basia
Aim:
Miniprep of overnight cultures
Method:
Miniprep Thermo Scientific according to the manual
Results:
Concentrations
| Sample | Concentration [ng/µL] |
| mut 1 | 530 |
| mut 2 | 595 |
| mut 3 | 221,7 |
| mut 4! | 63.5 |
| mut 5 | 360 |
| mut 6 | 350 |
| non mut 1 | 782,2 |
| non mut 2 | 523,3 |
| non mut 3 | 780 |
| non mut 4 | 651 |
| non mut 5 | 688 |
mut: cells that were always under arabinose and AID conditions
non mut: cells that are used as controls - never used arabinose and AID during cell culturing
send DNA from colonies after third round of phage display to sequencing
Investigators:
Basia/Chris
| sample | GATC number | Seq. Primer |
| mut 1 | AI2691 | GATC_Std_RPC |
| mut 2 | AI2692 | GATC_Std_RPC |
| mut 3 | AI2693 | GATC_Std_RPC |
| mut 4! | AI2699 | GATC_Std_RPC |
| mut 5 | AI2700 | GATC_Std_RPC |
| mut 6 | AI2717 | GATC_Std_RPC |
| non mut 1 | AI2694 | GATC_Std_RPC |
| non mut 2 | AI2695 | GATC_Std_RPC |
| non mut 3 | AI2696 | GATC_Std_RPC |
| non mut 4 | AI2697 | GATC_Std_RPC |
| non mut 5 | AI2698 | GATC_Std_RPC |
O/N culture of transformed E.coli with AID in RFP new standard cloning vector
Investigators: Rico, Sascha
Time: 2012-09-18
Method:
picking of clones, which show no red fluorescence indicating that the ligation was successful
2012-09-19
Checking AID induction test with SDS-PAGE gel
Investigators: Basia, Chris
There is no overexpressed lane that differs in the induced samples. Maybe the AID is low expressed because of the way the construct is built - 11 bp between the start codon and RBS instead of 7? The arrow shows where more less should be a band of AID.
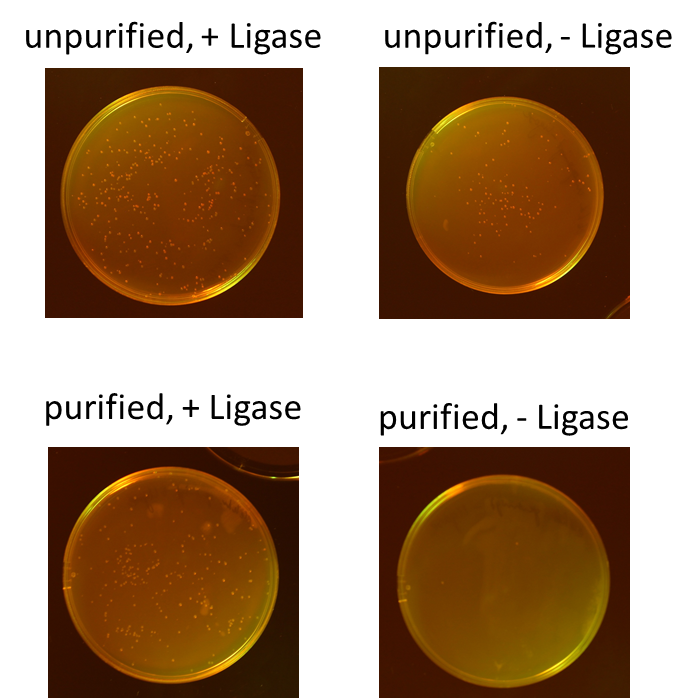
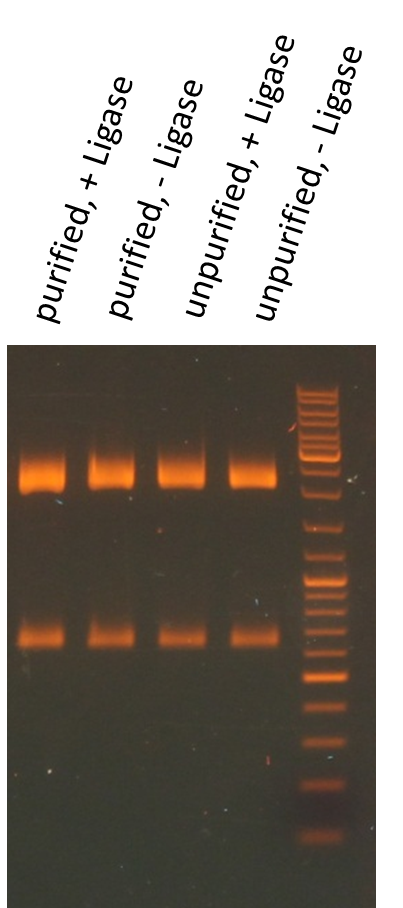
Miniprep of O/N cultures of RFP new standard cloning vector
Investigators: Rico, Sascha
Method: Miniprep kit from Thermo scientific
Results:
| sample | concentration in ng/µL |
| unpurified, + Ligase | 170.5 |
| unpurified, - Ligase | 174.8 |
| purified, + Ligase | 189.9 |
| purified, - Ligase | 179.1 |
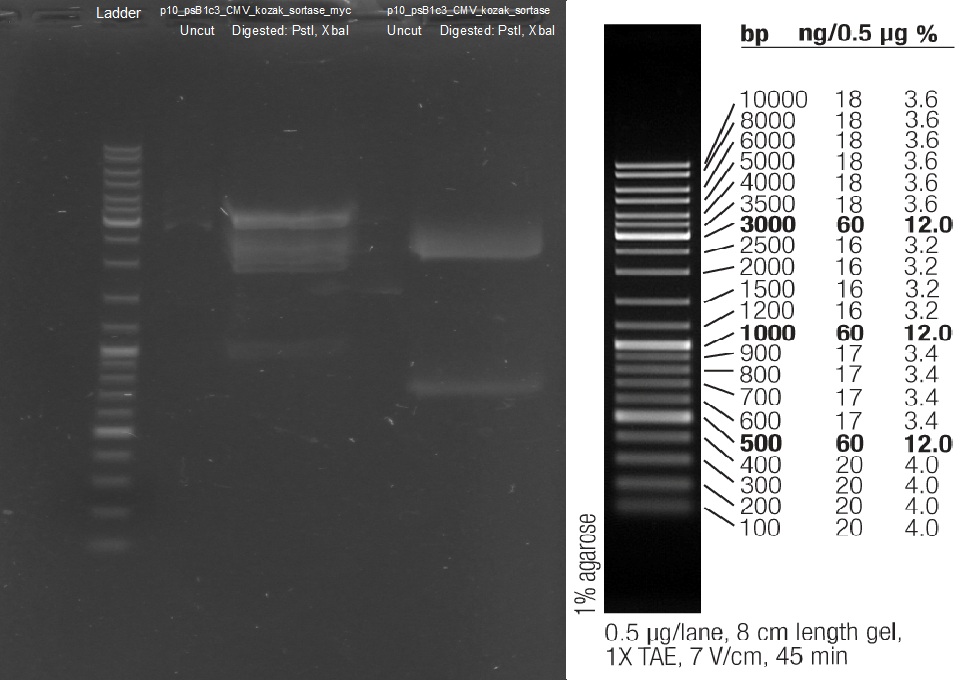
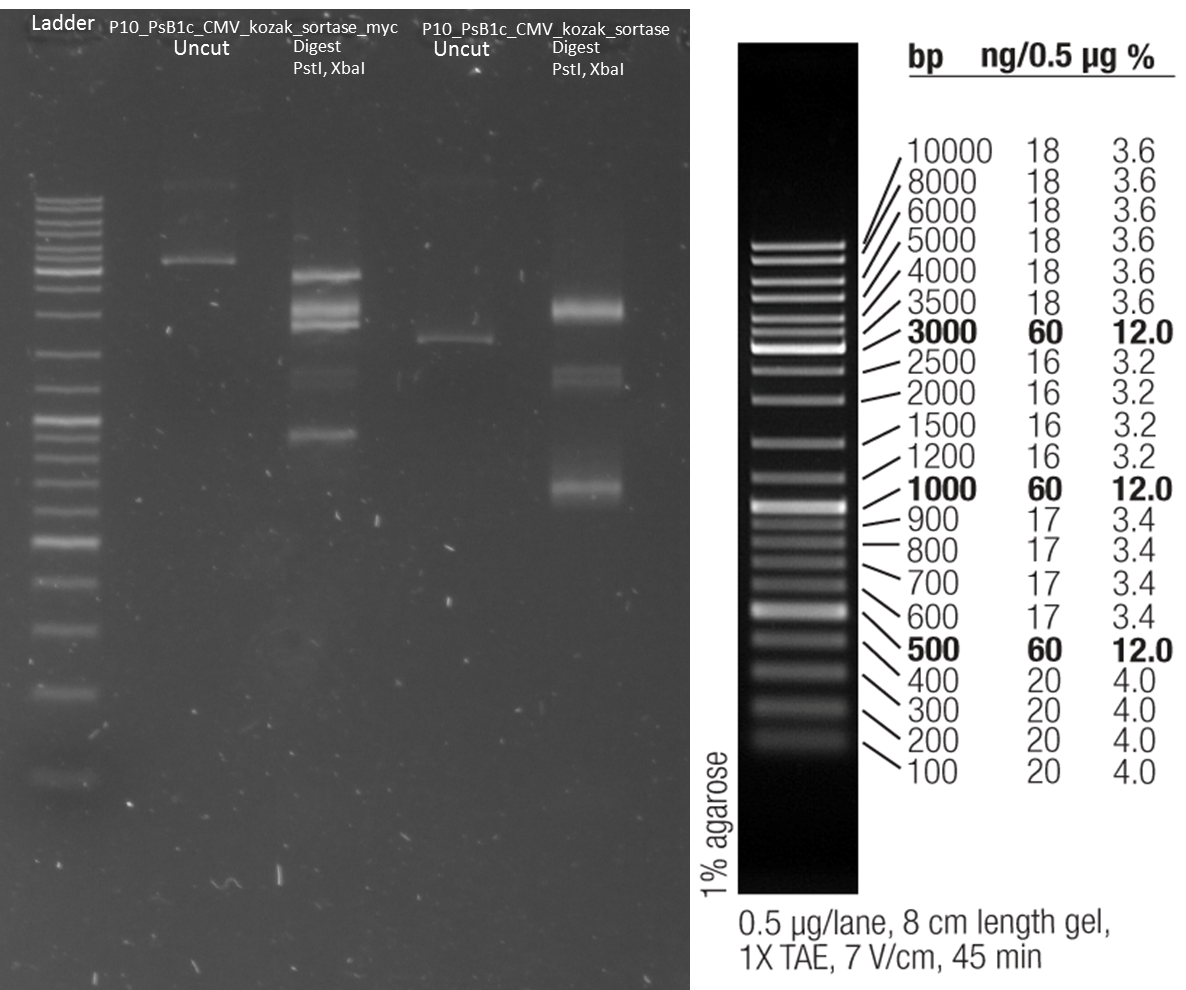
Test digestion of purified AID in new standard cloning vector
Investigators: Rico, Sascha
Results:
- all variants show the expected sizes of the digested fragments
Sending AID variants in new cloning vector and RFP cloning vector for sequencing
Investigators: Rico
Transfection of CHO cells with scFv-YFP, Nanobody-mCHerry and modified AID-eGFP construct for FACS
Investigators: Rico
Method: 2 µg DNA was used for transfection in 6 well plates
2012-09-20
Fluorescence microscopy of transfected CHO cells for FACS
Investigators: Rico
Results:
Cells shows a much higher red fluorescence (transfected with Nanobody-mCherry construct) than exprected
2012-09-21
Control of the sequencing results after phage display
Investigators: Basia
Results:
not a single mutation was introduced into the sequence. We think it is due to lack of AID expression in the pBAD vector.
2012-09-22
Selection of the antibodies from the supernatant after the Cre recombinase treatment via magnetic beads binding
Investigators: Basia
Materials:
Dyna Magnetic Beads bound with Streptavidin, biotinylated eGFP, supernatant after the CHO cell culturing, PBS buffer
Method: According to the Dyna beads Trial Kit Manual: 800pmol of biotinylated eGFP was used, 1ml of supernatant was added to the eGFP coupled with magnetic beads through streptavidin and biotin and incubated for 1hour with delicate mixing at room temperature. The samples were then resuspended in 100µl of loading buffer and loaded on the SDS protein gel.
Results:
Checking sequencing results for new standard
Investigators: Rico
Results:
all sequences show no mutation in the coding sequence, suffix and prefix
Antibody
2012-09-01
Glycerol stock and Mini Prep of Geneart construct, Venus, pAK100+scFv425 R H1, R H2, R L1, R L2, L1, L2
Investigator:Maria
Time: 2012-09-01 9:30 am
Materials: overnight cultures Geneart construct (4x), Venus (Tom), pAK100+scFv425 R H1, R H2, R L1, R L2, L1, L2 (Chris), Mini prep kit
Methods: according to manual
Results:
Venus: …. ng/µl
pAK100+scFv425 R H1: --- ng/µl
pAK100+scFv425 R H2: --- ng/µl<br<
pAK100+scFv425 R L1: --- ng/µl
pAK100+scFv425 R L2: --- ng/µl
pAK100+scFv425 L1: --- ng/µl
pAK100+scFv425 L2: --- ng/µl
Geneart construct 1: --- ng/µl
Geneart construct 2: --- ng/µl
Geneart construct 3: --- ng/µl
Geneart construct 4: --- ng/µl
+
Glycerole stocks of Geneart construct and pAK100+scFv425 R H1, R H2, R L1, R L2, L1, L2
Further tasks:
2012-09-03
cell culture;
Investigator:Kerstin/Stefan
Time: 2012-09-03 09:00 am
Topic: cell-culture
- seeding of zeocin selected CHO cells in 6-well + ibidi-dish for stable transfection and modified AID-GFP
- investigation of ordering phenolredfree medium
- searching fort he right version of imageJ
mutagenesis-PCR to remove PstI-sites off scFv-Tmd-EYFP-construct
Investigators: Sascha
Materials:
- Template: scFv-TEV-TMD-EYFP
- Phusion-polymerase
- 10x Phusion buffer HF
- dNTPs (10mM)
- Primer 1.RCF25 and 2.mut: RCF25 prefix and 1st mutation PstI in scFV (Tm=72°C)
- Primer 3.mut and 4.mut: 1st and 2nd mutation PstI in scFV (Tm= 72°C)
- Primer 5.mut and &.RCF25: 2nd mutation and RCF25 suffix (Tm= 72°C)
- Thermocycler
Methods: 50µl mix for primer 1 and 2
| reagent | volume [µL] |
| 10x Phusion HF buffer | 10 |
| dNTPs | 1 |
| Primer 1.RCF25 td> | 2,5 |
| Primer 2.mut: RCF25 prefix and 1st mutation PstI in scFV | 2,5 |
| DNA (scFv-TEV-TMD-EYFP | 1 |
| Phusion Polymerase | 0,5 |
| water | 32,5 |
Program for Primer 1+2
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 30 |
| annealing | 72 | 2 | 30 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
50µl mix for primer 3 and 4
| reagent | volume [µL] |
| 10x Phusion HF buffer | 10 |
| dNTPs | 1 |
| Primer 3.mut: 1st mutation PstI in scFV td> | 2,5 |
| Primer 4.mut: 1st mutation PstI in scFV | 2,5 |
| DNA (scFv-TEV-TMD-EYFP) | 1 |
| Phusion Polymerase | 0,5 |
| water | 32,5 |
Program for Primer 3+4
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 30 |
| annealing/elongation | 72 | 15 | 30 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
2012-09-04
cell culture;
Investigator:Kerstin/Stefan
Time: 2012-09-04 09:00 am
Topic: cell-culture
- stable transfection of clon 4, according to stable transfection protocoll
- transfection of AID-GFP in 6-well plate
- virus infection of CHO cells in 6-well plates and ibidi dishes
- evaluation of cell culture images
preparative digestion of pcdna5frt with NheI and ApaI
Investigator: Sascha
Materials:
- Fast Digest NheI
- Fast Digest ApaI
- 10x FD Green Buffer
- pcdna5frt (705,4ng/µl)
- sterile water
Method:
- 5µl pcdna5frt (705,4ng/µl)
- 2µl NheI
- 2µl ApaI
- 3µl 10x FD Green Buffer
- 18µl sterile water
- digestion for 2,h at 37°C
>Results:
Further Tasks:
- gelelectrophoresis
- gelextraction
dephosphorylating the vector pcDNA5FRT digested with NheI and ApaI
Investigator:Maria
Aim: dephosphorylating the digested pcDNA5-FRT to prevent re-ligation
Materials:
- Antarctic Phosphatase
- 10x Antarctic Phosphatase Reaction Buffer
- digested pcDNA5-FRT
Method:
- 3,3µl 10x Antarctic Phosphatase Reaction Buffer
- 1µl Antarctic Phosphatase
- incubation for 30min at 37°C to dephosphorylate 5´-ends of pcDNA5-FRT
- heat inactivation of Phosphatase for 5min at 65°C
- store at -20°C
Further Tasks:
- gelextraction
- ligation of pcDNA5-FRT dephosphorylated vector with geneart construct
PCR-Clean-Up of dephosphorylated vector
Investigator:Maria
Aim: cleaning of dephosphorylated vector
Materials:
- PCR-Clean-Up Kit
Method:
- according to manual
Results:
- concentration of cleaned dephosphorylated pcDNA5FRT = 60,4 ng/µl
colony-PCR of ligated pcDNA5-FRT_geneart-construct clones
Investigator: Maria
AIM: identifying correct ligated pcDNA5-FRT_geneart-constructs clones
Materials:
- Taq DNA-polymerase 1kb/1min
- 10x Taq DNA-polymerase buffer
- dNTPs (10mM)
- forward primer in C-terminus of CMV of pcDNA5-FRT-backbone: fp_qRT-CMV_rev
- reverse primer in C-terminus in mcherry: ps_mcherry_rev_BamHI * Thermocycler
Methods:
- 20µl colony-PCR mix for 20 clones:
Mastermix
| reagent | volume [µL] |
| 10x Standard Taq-DNA-Polymerase Buffer | 40 |
| dNTPs | 8 |
| Primer (Forward) | 8 |
| Primer (Reverse) | 8 |
| DNA | clone from plate |
| Taq Polymerase | 2 |
| water | 334 |
Primer forward: fp_qRT-CMV_rev(10µM)
Primer reverse: ps_mcherry_rev_BamHI (10µM)
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 95 | 300 | 1 |
| denaturation | 95 | 25 | 30 |
| annealing | 51 | 25 | 30 |
| elongation | 68 | 130 | 30 |
| final elongation | 86 | 300 | 1 |
| cooling | 8 | ∞ | 1 |
- clone-colonies were transferred to 20 µl colony-PCR mix, and onto LB-Amp plate, respectively
- LB-AMP-plate were incubated o.n. at 37°C
- negative control: clon #20 from religation-control (ligation-ratio 1:0) from 2012-09-03
further Taks:
- gelelectrophoresis
- picking positive clones for o.n.-culture
preparative gelelectrophoresis of digested scFv-TEV-TMD-EYFP
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
Method:
- 30µl digested clon4 geneart-nanobody-construct (451ng/µl)
- 1% agarose gel, 100ml
- 105V, 75min
Results:
- digested clon4 geneart-nanobody-construct (451ng/µl) with DNA @ 2029bp
Further Tasks:
- gel extraction
analytical gelelectrophoresis of three genes generated by mutagenesis-PCR from 2012-09-03 and assembled gene(primer 3+4) with gene(primer5+6)
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
- PCR-products
Method:
Each PCR-product: 10µl mix:
- 1µl DNA
- 1µl 10x FD Green
- 8µl steril water
- 1% agarose gel, 100ml
- 105V, 75min
Results:
Further Tasks:
- gel extraction
- ligation
mutagenesis-PCR to remove PstI-sites off scFv-Tmd-EYFP-construct
Investigators: Sascha
Materials:
- Template: scFv-TEV-TMD-EYFP
- Phusion-polymerase
- 10x Phusion buffer GC, 10x Phusion buffer GC
- DMSO
- dNTPs (10mM)
- Primer 1.RCF25 and 2.mut: RCF25 prefix and 1st mutation PstI in scFV (Tm=72°C)
- Primer 3.mut and 4.mut: 1st and 2nd mutation PstI in scFV (Tm= 72°C)
- Primer 5.mut and &.RCF25: 2nd mutation and RCF25 suffix (Tm= 72°C)
- Thermocycler
Methods: 50µl mixes for mutagenesis-PCR of gene(primer1+2)
| reagent | volume [µL]' | volume [µL] | volume [µL] | volume [µL] |
| 10x Phusion buffer | HF-Buffer 10 | HF-Buffer+DMSO 10 | GC-Buffer 10 | GC-Buffer + DMSO 10 |
| dNTPs | 1 | 1 | 1 | 1 |
| Primer 1.RCF25 prefix | 2,5 | 2,5 | 2,5 | 2,5 |
| Primer 2 1st mutation PstI in scFV 2,5 | 2,5 | 2,5 | 2,5 | 2,5 |
| DNA (scFv-TEV-TMD-EYFP) 1ng/µl | 1 | 1 | 1 | 1 |
| DMSO | 0 | 1,5 | 0 | 1,5 |
| Phusion Polymerase | 0,5 | 0,5 | 0,5 | 0,5 |
| water | 32,5 | 31 | 32,5 | 31 |
50µl mixes for mutagenesis-PCR of gene(primer(3+4)
| reagent | volume [µL] | volume [µL] | volume [µL] | volume [µL] |
| 10x Phusion buffer | HF-Buffer 10 | HF-Buffer+DMSO 10 | GC-Buffer 10 | GC-Buffer + DMSO 10 |
| dNTPs | 1 | 1 | 1 | 1 |
| Primer 3. 1st mutation in PstI in scFv 2,5 | 2,5 | 2,5 | 2,5 | 2,5 |
| Primer 4. 2nd mutation in PstI in scFv | 2,5 | 2,5 | 2,5 | 2,5 |
| DNA (scFv-TEV-TMD-EYFP) 1ng/µl | 1 | 1 | 1 | 1 |
| DMSO | 0 | 1,5 | 0 | 1,5 |
| Phusion Polymerase | 0,5 | 0,5 | 0,5 | 0,5 |
| water | 32,5 | 31 | 32,5 | 31 |
50µl mixes for mutagenesis-PCR of gene(primer(5+6)
| reagent | volume [µL] | volume [µL] | volume [µL] | volume [µL] |
| 10x Phusion buffer | HF-Buffer 10 | HF-Buffer+DMSO 10 | GC-Buffer 10 | GC-Buffer + DMSO 10 |
| dNTPs | 1 | 1 | 1 | 1 |
| Primer 5. 2nd mutation in PstI in scFv | 2,5 | 2,5 | 2,5 | 2,5 |
| Primer: RCF25 suffix | 2,5 | 2,5 | 2,5 | 2,5 |
| DNA (scFv-TEV-TMD-EYFP) 1ng/µl | 1 | 1 | 1 | 1 |
| DMSO | 0 | 1,5 | 0 | 1,5 |
| Phusion Polymerase | 0,5 | 0,5 | 0,5 | 0,5 |
| water | 32,5 | 31 | 32,5 | 31 |
Program
| step | Temperature [°C] | duration for gene(primer1+2) [s] | duration for gene(primer3+4) [s] | duration for gene(primer5+6) [s] | cycles |
| initial denaturation | 98 | 30 | 30 | 30 | 1 |
| denaturation | 98 | 8 | 8 | 8 | 5 |
| annealing | 71 | 10 | 10 | 10 | 5 |
| elongation | 72 | 5 | 8< /td> | 20 | 5 |
| denaturation | 98 | 8 | 8 | 8 | 25 |
| annealing | 72 | 8 | 15< /td> | 30 | 25 |
| final elongation | 72 | 600 | 600 | 600 | 1 |
| cooling | 8 | ∞ | ∞ | ∞ | 1 |
Further Taks:
- analytical gelelectrophoresis
- PCR-Clean up
Topic: analytical gelelectrophoresis mutagenesis -PCR to remove PstI-sites off scFv-Tmd-EYFP-construct gene(primer1+2), gene(primer3+4), gene(primer5+6)
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 90ml
- 105V
Results:
Further Tasks:
- PCR-Clean up
PCR-Clean up preparative gelelectrophoresis of scFv-TEV-TMD-EYFP-constructs after mutagenesis-PCR and concentration measurement
Investigator: Sascha
Materials:
- NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel)
Methods:
* according to CleanUp-protocol
Gene (primer1+2)= Gene (primer3+4)= Gene (primer5+6)=
assembly-PCR to of gene(primer1+2) with gene(primer(3+4)
Investigators: Sascha
Materials:
- Template: scFv-TEV-TMD-EYFP
- Phusion-polymerase
- 10x Phusion buffer HF
- DMSO
- dNTPs (10mM)
- Primer 1.RCF25 prefix (Tm=72°C)
- Primer6-.RCF25 suffix (Tm= 72°C)
- Thermocycler
Methods: 50µl mixes for assembly-PCR of gene(primer1+2) with gene(primer3+4)
| reagent | volume [µL]' | volume [µL] |
| 10x Phusion buffer | HF-Buffer 10 | HF-Buffer+DMSO 10 |
| dNTPs | 1 | 1 |
| Primer 1.RCF25 prefix | 2,5 | 2,5 |
| Primer6-.RCF25 suffix | 2,5 | 2,5 |
| template gene(primer1+2) 1ng/µl | 2 | 2 |
| template gene(primer3+4) 0,25ng/µl | 1 | 1 |
| DMSO | 0 | 1,5 |
| Phusion Polymerase | 0,5 | 0,5 |
| water | 32,5 | 31 |
preparative gelelectrophoresis of assembled gene(primer1+2) with gene(primer3+4) to gene12
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
Method:
- 48µl of assembled gene12
* 1% agarose gel, 100ml
- 105V, 75min
Results:
- assembly was successful; DNA @ bp
Further Tasks:
- gelextraction
gel extraction of assembled gene(primer1+2) with gene(primer3+4) to gene12 and concentration measurement
Investigators: Sascha
Materials:
- Gel-Clean-Up Kit
Method:
- according to manual
Results:
- gene12= -- ng/µl
Further tasks:
- assembly of gene12 to gene(primer5+6)
assembly-PCR of gene12 and gene(primer5+6) to gene4
Investigators: Sascha
Materials:
- Templates:gene12 and gene(primer5+6)
- Phusion-polymerase
- 10x Phusion buffer GC, 10x Phusion buffer GC
- DMSO
- dNTPs (10mM)
- Primer: 1.RCF25 RCF25 prefix (Tm=72°C)
- Primer: RCF25 suffix (Tm= 72°C)
- Thermocycler
Methods: 50µl mixes for assembly-PCR of gene12 with gene(primer5+6)
| reagent | volume [µL]' | volume [µL] | volume [µL] | volume [µL] |
| 10x Phusion buffer | GC -Buffer 10 | GC -Buffer+DMSO 10 | HF -Buffer 10 | HF -Buffer + DMSO 10 |
| dNTPs | 1 | 1 | 1 | 1 |
| Primer 1.RCF25 prefix | 2,5 | 2,5 | 2,5 | 2,5 |
| Primer 2 1st mutation PstI in scFV 2,5 | 2,5 | 2,5 | 2,5 | 2,5 |
| gene12 (2,4ng/µl) | 1 | 1 | 1 | 1 |
| gene(primer5+6) (1ng/µl) | 1 | 1 | 1 | 1 |
| DMSO | 0 | 1,5 | 0 | 1,5 |
| Phusion Polymerase | 0,5 | 0,5 | 0,5 | 0,5 |
| water | 32,5 | 31 | 32,5 | 31 |
Program
| step | Temperature [°C] | duration for gene4 GC-buffer [s] | duration for gene4 GC-buffer + DMSO [s] | duration for gene4 HF-buffer [s] | duration for gene4 HF-buffer + DMSO [s] | cycles |
| initial denaturation | 98 | 30 | 30 | 30 | 30 | 1 |
| denaturation | 98 | 8 | 8 | 8 | 8 | 5 |
| annealing | 71 | 10 | 10 | 10 | 10 | 5 |
| elongation | 72 | 20 | 20 | 20 | 20 | 5 |
| denaturation | 98 | 8 | 8 | 8 | 8 | 25 |
| annealing | 72 | 35 | 35 | 35 | 35 | 25 |
| final elongation | 72 | 600 | 600 | 600 | 600 | 1 |
| cooling | 8 | ∞ | ∞ | ∞ | ∞ | 1 |
Further Taks:
- analytical gel electrophoresis
- PCR-Clean up
Topic: analytical gelelectrophoresis of assembled gene4
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 90ml
- 2µl gene4, 1µl FD Green, 8µl water
- 105V,80min
Results:
Further Tasks:
- preparative gel electrophoresis of gene4
- gel extraction of gene4
2012-09-05
cell culture;
Investigator:Kerstin/Stefan
Time: 2012-09-05 10:00 am
Topic: cell-culture
- change of medium of transfected (stable transfection clon 4 and AID-GFP ) cells (04.09.2012)
- passaging of CHO’s
- seeding of 2x 6-wells with each 2*10^5 cells/well
- seeding of one ibidi dish with 5*10^4 cells/well
- talking with Daniel to check the FACS options at the junior group of potsdam university
- investigation about Geniticin and possible resistance genes
analytical gelelectrophoresis of colony PCR with 20 clones of ligated pcDNA5-FRT-geneart construct
Investigator: Maria
Aim: analytical gelelectrophoresis to check successful ligation (geneart construct and pcDNA5-FRT vector)
Materials:
- agarose
- 1xTAE-buffer
- 10xFD FDGreen
Method:
- 3µl 10x FD Green, 15µl of each PCR product
- 1% agarose gel, 100ml
- 70 min at 120V
Results:
- no precise bands
Further Tasks:
- re-doing colony PCR
preparative gelelectrophoresis of assembled gene4
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
Method:
- 48µl of assembled gene4
* 1% agarose gel, 100ml
- 105V, 85min
Results:
- assembly was successful; DNA @ bp
Further Tasks:
- gelextraction
gel extraction of assembled gene4
Investigators: Sascha
Materials:
- Gel-Clean-Up Kit
Method:
- according to manual
Results:
- gene4= -- ng/µl
Further tasks:
gel extraction of assembled gene(primer1+2) with gene(primer3+4) to gene12 and concentration measurement
Investigators: Sascha
Materials:
- Gel-Clean-Up Kit
Method:
- according to manual
Results:
- gene12= -- ng/µl
Further tasks:
- assembly of gene12 to gene(primer5+6)
PCR of scFv-only with RE-site for biobricks
Investigators: Sascha
Materials:
- Templates:gene12 and gene(primer5+6)
- Phusion-polymerase
- 10x Phusion buffer GC, 10x Phusion buffer GC
- DMSO
- dNTPs (10mM)
- Primer scFv: Primer 7.RFC25 prefix and 8.RCF25 suffix
- Thermocycler
Methods: 50µl mixes
| reagent | volume [µL]' | volume [µL] | volume [µL] | volume [µL] |
| 10x Phusion buffer | GC -Buffer 10 | GC -Buffer+DMSO 10 | HF -Buffer 10 | HF -Buffer + DMSO 10 |
| dNTPs | 1 | 1 | 1 | 1 |
| Primer 7.Rcf25 prefix | 2,5 | 2,5 | 2,5 | 2,5 |
| Primer 8.RCF25 suffix | 2,5 | 2,5 | 2,5 | 2,5 |
| gene12 (1ng/µl) | 10 | 10 | 10 | 10 |
| DMSO | 0 | 1,5 | 0 | 1,5 |
| Phusion Polymerase | 0,5 | 0,5 | 0,5 | 0,5 |
| water | 33,5 | 32 | 33,5 | 32 |
Program
| step | Temperature [°C] | duration for scFv-only GC-buffer [s] | duration for scFv-only GC-buffer + DMSO [s] | duration for scFv-only HF-buffer [s] | duration for scFv-only HF-buffer + DMSO [s] | cycles |
| initial denaturation | 98 | 30 | 30 | 30 | 30 | 1 |
| denaturation | 98 | 8 | 8 | 8 | 8 | 5 |
| annealing | 72 | 15 | 15 | 15 | 15 | 5 |
| final elongation | 72 | 600 | 600 | 600 | 600 | 1 |
| cooling | 8 | ∞ | ∞ | ∞ | ∞ | 1 |
Further Taks:
- analytical gel electrophoresis
- PCR-Clean up
2012-09-06
cell culture;
Investigator:Kerstin/Stefan
Time: 2012-09-06 10:00 am
Topic: cell-culture
- stable transfected CHO’s (clone 4) were diluted from 6-well in 75cm^2 flasks
- capturing images of virus infected cells
- talking with Daniel concerning use of FACS
- planing of AK verification at the Cellmembrane ==> anti flag tag
colony-PCR of ligated pcDNA5-FRT_geneart-construct clones from 05.09.12
Investigator: Maria
AIM: identifying correct ligated pcDNA5-FRT_geneart-constructs clones
Materials:
- 20µl colony-PCR mix for 32 clones:
- Taq DNA-polymerase 1kb/1min
- 10x Taq DNA-polymerase buffer
- dNTPs (10mM)
- forward primer in C-terminus of CMV of pcDNA5-FRT-backbone: fp_qRT-CMV_rev (Tm=55°C)
- reverse primer in C-terminus in mcherry: ps_mcherry_rev_BamHI (Tm= 59,31°C)
- Thermocycler
Methods:
Mastermix
| reagent | volume [µL] |
| 10x Standard Taq-DNA-Polymerase Buffer | 64 |
| dNTPs | 12,8 |
| Primer (Forward) | 12,8 |
| Primer (Reverse) | 12,8 |
| DNA | clone from plate |
| Taq Polymerase | 3,2 |
| water | 534,4 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 95 | 300 | 1 |
| denaturation | 95 | 25 | 30 |
| annealing | 51 | 25 | 30 |
| elongation | 68 | 130 | 30 |
| final elongation | 86 | 300 | 1 |
| cooling | 8 | ∞ | 1 |
- clone-colonies were transferred to 20µl colony-PCR mix, and onto LB-Amp plate, respectively
- LB-AMP-plate were incubated o.n. at 37°C
- negative control: clone #1 from religation-control (ligation-ratio 1:0) from 2012-09-05, clone 2 from transformation control of new competent E.coli Xl 1 blue with pcDNA5FRT
further Taks:
- gelelectrophoresis
- picking positive clones for o.n. culture
- o.n. culture of positive clones
- test digestion
- o.n. culture for endotoxin free preparation
analytical gelelectrophoresis of colony PCR with 32 clones of ligated pcDNA5-FRT-geneart construct, circular vector and religated vector
Investigator: Maria
Aim: analytical gelelectrophoresis to check successful ligation (geneart construct and pcDNA5-FRT vector)
Materials:
- agarose
- 1xTAE-buffer
- 10xFD FDGreen
Method:
- 3µl 10x FD Green, 15µl of each PCR product
- 1% agarose gel, 100ml
- 120V
Results:
- no precise bands
Further Tasks:
- picking promising clones
- o.n. culture of promising clones
Transformation of pcDNA5FRT into new XL1-blue competent E. coli cells
Investigators: Maria
Materials:
- Bunsen Burner, Agar Plate with Ampicillin, 37 °C thermo mixer, centrifuge,
- pcDNA5FRT
- icebox
- new competent E. coli cells (XL 1)
Method:
- according to manual
- 20µl of resuspended cell-suspension were plated on a LB-Amp-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones
Topic: analytical gelelectrophoresis of assembled gene4
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 90ml
- 2µl ScFv of each PCR-mix, 1µl FD Green, 8µl water
- 105V,80min
Results:
Further Tasks:
- preparative digestion of scFv-only_delta PstI, scFv-construct_delta PstI, BBA_K404316
preparative digestion of scFv-only_delta PstI, scFv-construct_delta PstI, BBA_K404316 with NgoMIV and AgeI
Investigator: Sascha
Materials:
- Fast Digest NgoMIV
- Fast Digest AgeI
- 10x FD Green Buffer
- pcdna5frt (705,4ng/µl)
- sterile water
Method:
- approximately 500ng of DNA (scFv-only_delta PstI, scFv-construct_delta PstI, BBA_K404316)
- 1µl NgoMIV
- 1µl AgeI
- 3µl 10x FD Green Buffer
- sterile water ad 30µl
- digestion for 1,5h at 37°C
>Results:
- suboptimal restrictions enzymes
Further Tasks:
- new digestion with XbaI and AgeI
- new assembly to receive more DNA
PCR to generate biobrick-ready genes from geneart-nanobody construct
Investigators: Sascha
Materials:
- Templates: geneart-nanobody construct
- Phusion-polymerase
- 10x Phusion buffer GC, 10x Phusion buffer HF
- DMSO
- dNTPs (10mM)
- Primer 1.1 and 1.2: Signal peptide-Nanobody-Fc
- Primer 2.1 and 2.2: TEV-LoxP-TMD-mcherry-LoxP
- Primer 3.1 and 3.2: Nanobody
- Primer 4.1 and 4.2(=1.2): Fc
- Primer5.1 and 5.2: TMD-mcherry
- Thermocycler
Methods: 50µl mix for every biobrick-ready gene, PCR-mix applies for all genes
| reagent | volume [µL]' | volume [µL] | volume [µL] | volume [µL] |
| 10x Phusion buffer | GC -Buffer 10 | GC -Buffer+DMSO 10 | HF -Buffer 10 | HF -Buffer + DMSO 10 |
| dNTPs | 1 | 1 | 1 | 1 |
| Primer 1.1: Signal peptide-Nanobody-Fc; Primer 2.1: TEV-LoxP-TMD-mcherry-LoxP ; Primer 3.1:Nanobody; Primer 4.1: Fc; Primer5.1: TMD-mcherry | 2,5 | 2,5 | 2,5 | 2,5 |
| Primer 1.2: Signal peptide-Nanobody-Fc; Primer 2.2: TEV-LoxP-TMD-mcherry-LoxP ; Primer 3.2: Nanobody; Primer 4.2(=1.2): Fc; Primer 5.2: TMD-mcherry | 2,5 | 2,5 | 2,5 | 2,5 |
| geneart-nanobody construct (1ng/µl)td> | 1 | 1 | 1 | 1 |
| DMSO | 0 | 1,5 | 0 | 1,5 |
| Phusion Polymerase | 0,5 | 0,5 | 0,5 | 0,5 |
| water | 32,5 | 31 | 32,5 | 31 |
Program for gene2,4,5
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| elongation | 72 | 14 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing/elongation | 72 | 15 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Program for gene1
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| elongation | 72 | 18 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing/elongation | 72 | 20 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Program for gene3
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| elongation | 72 | 6 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing/elongation | 72 | 8 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: analytical gelelectrophoresis of assembled gene4
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 90ml
- 2µl of each PCR-mix, 1µl FD Green, 8µl water
- 105V,80min
Results:
- unspecific and incorrect DNA bands
Further Tasks:
- preperative digestion of genart-nanobody-construct to facilitate better annealing of primer to geneart-nanobody construct
2012-09-07
cell culture;
Investigator:Kerstin/Stefan
Time: 2012-09-07 10:00 am
Topic: cell-culture
- passaging of CHO cells w/ zeocin and w/o zeocin
- capturing images of AID-GFP
picking promising clones of ligated pcDNA5-FRT-geneart construct and of new competent cells with pcDNA5FRT, inoculation of o.n.-culture
Investigators: Maria
Materials:
- 11x 15 ml inoculation tubes
- LB-Medium
- Ampicilin
- bunsen burner
Method:
- 10x o.n.-culture of promising pcDNA5-FRT-geneart construct clones
- 1x o.n.-culture of control for new competent cells
- each o.n.-culture 5ml LB-Medium + 5µl Ampicillin
- incubation o.n. at 37°C
further Tasks:
- miniprep of o.n.-cultures
preparative digestion of geneart-nanobody-construct with NheI and ApaI
Investigator: Sascha
Materials:
- Fast Digest NheI
- Fast Digest ApaI
- 10x FD Green Buffer
- geneart-nanobody-construct (450ng/µl)
- sterile water
Method:
- 3µl geneart-nanobody-construct (450ng/µl)
- 1µl NgoMIV
- 1µl AgeI
- 3µl 10x FD Green Buffer
- sterile water ad 30µl
- digestion for 1,5h at 37°C
>Results:
- suboptimal restrictions enzymes
Further Tasks:
- new digestion with XbaI and AgeI
new assembly to receive more DNA
2012-09-08
Glycerol stock and Mini Prep of promising pcDNA5-FRT-geneart construct clones and new competent cells with pcDNA5FRT
Investigator: Maria
Materials: overnight cultures pcDNA5-FRT-geneart construct clones, control of new competent cells with pcDNA5FRT, Mini prep kit
Methods: according to manual
Results:
Clon 4: 589,7 ng/µl
Clon 5: 628 ng/µl<br<
Clon 7: 568,7 ng/µl
Clon 10: 613,4 ng/µl
Clon 11: 573,1 ng/µl
Clon 12: 682,7 ng/µl
Clon 14: 648,1 ng/µl
Clon 16: 615,8 ng/µl
Clon 22: 653,4 ng/µl
Clon 27: 602,5 ng/µl
Clon 29: 613,9 ng/µl
Controle pcDNA5FRT: 205,3 ng/µl
Glycerole stocks of pcDNA5-FRT-geneart construct clones
Further tasks: analytical digestion, analytical gelelectrophoresis, stable transfection of CHO cells
analytical digestion of ligated pcDNA5-FRT-geneart construct and pcDNA5FRT control
Investigator: Maria
Materials:
- all 10 prepared pcDNA5-FRT-geneart construct clones
- pcDNA5FRT vector control for new competent cells
- FastDigest ScaI and ApaI
- 10x FD Green Buffer
- sterile water
Method:
- 10µl mix: 1µl ScaI for ligated construct, 1µl ApaI for vector, 1µl 10x FD Green Buffer, 1µl of each clone (approximately 200ng DNA), sterile water add to 10µl
- incubation at 37°C for 30 min
Further tasks:
- analytical gelelectrophoresis
Topic: gelelectrophoresis of analytical digested pcDNA5-FRT-geneart construct and pcDNA5FRT control
Investigator: Maria
Aim: checking plasmid-size after ligation of pcDNA5-FRT-geneart construct and pcDNA5FRT control in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 100ml
- 120 V
Results:
- ligation successfully
Further Tasks:
- o.n. (50 µl) of positive clones 5, 14, 16 and 29 for endotoxin free prep
2012-09-09
Endotoxin free preparation of clones 5, 14,16 and 29
Investigator:Maria
Materials: endotoxin free Mediprep kit, overnight culture CMV-Venus
Methods: according to manual
Results: <b/>
clone 5: 6909,5 ng/µl
clone 14: 6243,6 ng/µl
clone 16: 3837,3 ng/µl
clone 29: 2454,8 ng/µl
Further tasks: transient and stable transfection of CHO cells
picking clones of scFv only and scFv construct ligated into pSB1C3 for overnight culture (vector and construct digested with AgeI and NgoMVI)
Investigator: Maria
Materials:
- 15 ml inoculation tubes
- LB-Medium
- Chloramphenicol
- bunsen burner
Method:
- 8x o.n.-culture of scFv only
- 8x o.n.-culture of scFv construct
- each o.n.-culture 5ml LB-Medium + 5µl Cm
- incubation o.n. at 37°C
further Tasks:
- miniprep of o.n.-cultures
cell culture
Investigator:Kerstin
Topic: cell-culture
- seeding CHO cells in 6-well plate for stable transfection
preparative gelelectrophoresis of digested nanobody-construct , digested psB1C3-vector and analytical electrophoresis of wt-AID with thiophosphate overhang
Investigator: Rico,Sascha
Materials:
- agarose
- digested DNA( nanobody-construct , psB1C3-vector) and wt-AID with thiophosphate overhang
- 1xTAE-buffer
- digested DNA
Method:
- 38µl of digestd nanobody-construct(NheI, ApaI)
- 28µl of digested psB1C3
- 2x 2µl of wt-Thio-AID
- 1% agarose gel, 100ml
- 105V, 85min
Results:
Further Tasks:
- gelextraction
gel extraction of digested nanobody-construct , digested psB1C3-vector
Investigators: Sascha
Materials:
- Gel-Clean-Up Kit
Method:
- according to manual
Results:
- nanobody-construct(NheI + ApaI)= -- ng/µl
- psB1C3 (XbaI + PstI)
Further tasks:
- PCR-clean up of wt-Thio-Aid
PCR to generate biobrick-ready genes 2,4,5 from digested geneart-nanobody construct
Investigators: Sascha
Materials:
- Templates: digested geneart-nanobody construct
- Phusion-polymerase
- 10x Phusion buffer GC, 10x Phusion buffer HF
- DMSO
- dNTPs (10mM)
- Primer 2.1 and 2.2: TEV-LoxP-TMD-mcherry-LoxP
- Primer 4.1 and 4.2(=1.2): Fc
- Primer5.1 and 5.2: TMD-mcherry
- Thermocycler
Methods: PCR mastermixes ( for 50µl single mix for every biobrick-ready gene, PCR-mix applies for all genes)
| reagent | volume [µL]' | volume [µL] | volume [µL] | volume [µL] |
| 10x Phusion buffer | GC -Buffer 30 | GC -Buffer+DMSO 30 | HF -Buffer310 | HF -Buffer + DMSO 30 |
| dNTPs | 3 | 3 | 3 | 3 |
| Mastermix for primer: Primer 2.1: TEV-LoxP-TMD-mcherry-LoxP ; Primer 4.1: Fc; Primer5.1: TMD-mcherry | 10 | 10 | 10 | 10 |
| Mastermix for primer: Primer 2.2: TEV-LoxP-TMD-mcherry-LoxP ; Primer 4.2(=1.2): Fc; Primer 5.2: TMD-mcherry | 10 | 10 | 10 | 10 |
| geneart-nanobody construct (1ng/µl)td> | 3 | 3 | 3 | 3 |
| DMSO | 0 | 4,5 | 0 | 4,5 |
| Phusion Polymerase | 1,5 | 1,5 | 1,5 | 1,5 |
| water | 97,5 | 93 | 97,5 | 93 |
Program for gene2,4,5
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| elongation | 72 | 14 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing/elongation | 72 | 15 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
PCR to generate biobrick-ready genes 1 from digested geneart-nanobody construct
Investigators: Sascha
Materials:
- Templates: digested geneart-nanobody construct
- Phusion-polymerase
- 10x Phusion buffer GC, 10x Phusion buffer HF
- DMSO
- dNTPs (10mM)
- Primer 1.1 and 1.2: Signal peptide-Nanobody-Fc
- Thermocycler
Methods: 50µl mix for every biobrick-ready gene, PCR-mix applies for all genes
| reagent | volume [µL]' | volume [µL] | volume [µL] | volume [µL] |
| 10x Phusion buffer | GC -Buffer 10 | GC -Buffer+DMSO 10 | HF -Buffer 10 | HF -Buffer + DMSO 10 |
| dNTPs | 1 | 1 | 1 | 1 |
| Primer 1.1: Signal peptide-Nanobody-Fc | 2,5 | 2,5 | 2,5 | 2,5 |
| Primer 1.2: Signal peptide-Nanobody-Fc | 2,5 | 2,5 | 2,5 | 2,5 |
| geneart-nanobody construct (1ng/µl)td> | 3 | 3 | 3 | 3 |
| DMSO | 0 | 1,5 | 0 | 1,5 |
| Phusion Polymerase | 0,5 | 0,5 | 0,5 | 0,5 |
| water | 32,5 | 31 | 32,5 | 31 |
Program for gene1
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| elongation | 72 | 18 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing/elongation | 72 | 20 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: analytical gelelectrophoresis of amplified gene2,4,5,1
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 90ml
- 2µl of each PCR-mix, 1µl FD Green, 8µl water
- 110V,75min
Results:
Further Tasks:
- furhter amplification of genes 1, 2, 4
2012-09-10
Mini Prep of pcDNA5-FRT-scFv only and -scFv construct clones
Investigator:Maria
Materials: overnight cultures pcDNA5-FRT-scFv only /-scFv construct clones, Mini prep kit
Methods: according to manual
Results:
scFv only
Clon I 1: 54,2 ng/µl
Clon I 2: 70,1 ng/µl<br<
Clon I 3: 60,6 ng/µl
Clon I 4: 168,3 ng/µl
Clon II 1: 138,6 ng/µl
Clon II 2: 160,2 ng/µl
Clon II 3: 149,8 ng/µl
Clon II 4: 138,8 ng/µl
Clon III 1: 160,5 ng/µl
Clon III 2: 170,5 ng/µl
Clon III 3: 127,9 ng/µl
Clon III 4: 170,9 ng/µl
Clon VI 1: 155,4 ng/µl
Clon VI 2: 116 ng/µl
Clon VI 3: 128,1 ng/µl
Clon VI 4: 96,4 ng/µl
Further tasks: analytical digestion, analytical gelelectrophoresis
analytical digestion of ligated pcDNA5-FRT-scFv only and scFv construct
Investigators:Maria
Materials:
- all 16 prepared pcDNA5-FRT-scFv only and –scFv construct clones
- FastDigest ScaI and AcuI
- 10x FD Green Buffer
- sterile water
Method:
- 10µl mix: 1µl AcuI, 1µl 10x FD Green Buffer, 2µl or 1µl of each clone respectively (approximately 150 ng DNA), sterile water add to 10µl
- incubation at 37°C for 30 min
further tasks:
- analytical gelelectrophoresis
Topic: gelelectrophoresis of analytical digested pcDNA5-FRT-scFv only and –scFv construct
Investigators:Maria
Aim: checking plasmid-size after ligation of pcDNA5-FRT-scFv only and -scFv construct in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 100ml
- 120 V
Results:
- ligation not successful
Further Tasks:
- Redoing ligation
Topic: taking cre recombinase (pBS185 CMV-Cre) into culture
Investigators:Maria
Aim: taking cre recombinase into culture
Materials:
- cre recombinase (addgene: pBS185)
- Agar plate with ampecillin
Method:
- streaking cre recombinase onto agar ampicillin plate
Results:
- cre recombinase clones for o.n. culture
Further Tasks:
- o.n. culture of cre recombinase
cell culture
Investigator:Kerstin
Topic: cell-culture
- stable transfection of CHO cells with Nanobody construct (Clone 29) in CHO cells
- passaging of CHO cells w/ zeocin and w/o zeocin
- taking HT1080 cells into culture
- starting to test sensitivity of CHO Flp-In cells to G418: cells were seeded in 24-well plate and incubated with different concentrations of G418 (0,1 mg/ml - 1,5mg/ml in 15 steps) for 10 days
PCR to generate biobrick-ready genes from geneart-nanobody construct
Investigators: Sascha
Materials:
- Templates: geneart-nanobody construct
- Phusion-polymerase
- 10x Phusion buffer HF
- DMSO
- dNTPs (10mM)
- Primer 1.1 and 1.2: Signal peptide-Nanobody-Fc
- Primer 2.1 and 2.2: TEV-LoxP-TMD-mcherry-LoxP
- Primer 3.1 and 3.2: Nanobody
- Primer 4.1 and 4.2(=1.2): Fc
- Primer5.1 and 5.2: TMD-mcherry
- Thermocycler
Methods: 50µl mix for every biobrick-ready gene, PCR-mix applies for all genes
| reagent | volume [µL]' |
| 10x Phusion HF-buffer | 10 |
| dNTPs | 1 |
| Primer 1.1: Signal peptide-Nanobody-Fc; Primer 2.1: TEV-LoxP-TMD-mcherry-LoxP ; Primer 3.1:Nanobody; Primer 4.1: Fc; Primer5.1: TMD-mcherry | 2,5 |
| Primer 1.2: Signal peptide-Nanobody-Fc; Primer 2.2: TEV-LoxP-TMD-mcherry-LoxP ; Primer 3.2: Nanobody; Primer 4.2(=1.2): Fc; Primer 5.2: TMD-mcherry | 2,5 |
| geneart-nanobody construct (2ng/µl) | 2 |
| Phusion Polymerase | 0,5 |
| water | 32,5 |
Program for gene2,4,5
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| elongation | 72 | 16 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing/elongation | 72 | 16 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
</table>
Program for gene1
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| elongation | 72 | 18 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing/elongation | 72 | 20 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Program for gene3
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| elongation | 72 | 6 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing/elongation | 72 | 8 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: analytical gelelectrophoresis of amplified gene1,2,3,4,5
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 90ml
- 2µl of each PCR-mix, 1µl FD Green, 8µl water
- 110V,80min
Results:
- only genes 3 (~350bp) and 5(~800bp) show DNA at desired bp-size
Further Tasks:
- further amplification of genes 1, 2, 4
2012-09-11
assembly PCR of mutated scFv without PstI -TEV-TMD-EYFP construct
Investigator: Maria
AIM: assembly of scFV-TEV-TMD-EGFP without PstI restriction sites
Materials:
- Template mutated scFv and TEV-TMD-EGFP
- Phusion-polymerase
- 10x Phusion buffer GC
- dNTPs (10mM)
- forward primer: small-1-fw-prefix25 (Tm=72°C)
- reverse primer: small-6-rw-suffix-25 (Tm= 72°C)
- DSMSO
- Thermocycler
Methods:
Mastermix for triplex batch
| reagent | volume [µL] |
| 10x Phusion GC buffer | 30 |
| dNTPs | 3 |
| Primer (Forward) | 7,5 |
| Primer (Reverse) | 7,5 |
| template 1 | 3 |
| template 2 | 3 |
| DMSO | 4,5 |
| Phusion Polymerase | 1,5 |
| water | 87 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| annealing + elongation | 72 | 30 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing | 72 | 35 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Further Taks:
- analytical gelelectrophoresis
Topic: analytical gelelectrophoresis of PCR assembly product scFv-TEV-TMD-EGFP
Investigator: Maria
Aim: checking assembled scFv-TEV-TMD-EGFP PCR result in 1% agarose gel
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 100ml
- 120 V
Results:
- assembly PCR successfully
Further Tasks:
- preparative digestion with AgeI and XbaI
- preparative gelelectrophoresis
- gel extraction
assembly PCR of mutated scFv without PstI and TEV-TMD-EYFP construct
Investigator: Maria
AIM: assembly of scFV-TEV-TMD-EGFP without PstI restriction sites in 4 batches
Materials:
- Template mutated scFv and TEV-TMD-EGFP
- Phusion-polymerase
- 10x Phusion buffer GC and 10x Phusion HD buffer
- dNTPs (10mM)
- forward primer: small-1-fw-prefix25 (Tm=72°C)
- reverse primer: small-6-rw-suffix-25 (Tm= 72°C)
- DSMSO
- Thermocycler
Methods:
Mastermix 1, 2x
| reagent | volume [µL] |
| 10x Phusion GC buffer | 20 |
| dNTPs | 2 |
| Primer (Forward) | 5 |
| Primer (Reverse) | 5 |
| template 1 | 2 |
| template 2 | 2 |
| DMSO | - |
| Phusion Polymerase | 1 |
| water | 63 |
Mastermix 2, 2x
| reagent | volume [µL] |
| 10x Phusion GC buffer | 20 |
| dNTPs | 2 |
| Primer (Forward) | 5 |
| Primer (Reverse) | 5 |
| template 1 | 2 |
| template 2 | 2 |
| DMSO | 3 |
| Phusion Polymerase | 1 |
| water | 60 |
Mastermix 3, 2x
| reagent | volume [µL] |
| 10x Phusion HC buffer | 20 |
| dNTPs | 2 |
| Primer (Forward) | 5 |
| Primer (Reverse) | 5 |
| template 1 | 2 |
| template 2 | 2 |
| DMSO | - |
| Phusion Polymerase | 1 |
| water | 63 |
Mastermix 4, 2x
| reagent | volume [µL] |
| 10x Phusion HC buffer | 20 |
| dNTPs | 2 |
| Primer (Forward) | 5 |
| Primer (Reverse) | 5 |
| template 1 | 2 |
| template 2 | 2 |
| DMSO | 3 |
| Phusion Polymerase | 1 |
| water | 60 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| annealing + elongation | 72 | 30 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing | 72 | 35 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Further Taks:
- analytical gelelectrophoresis
taking cre recombinase (pBS185 CMV-Cre) into culture for single clone picking
Investigators:Maria
Aim: taking cre recombinase into culture
Materials:
- cre recombinase (addgene: pBS185)
- Agar plate with ampicillin
Method:
- streaking cre recombinase onto agar ampicillin plate, single colonies wanted
Results:
- cre recombinase clones for o.n. culture
Further Tasks:
- o.n. culture of cre recombinase
Transformation of eGFP ( BBa_404316) into XL1-blue competent E. coli cells
Investigators: Maria
Materials:
- Bunsen Burner, Agar Plate with Ampicillin, 37 °C thermo mixer, centrifuge,
- BBa_404316
- icebox
- new competent E. coli cells (XL 1)
Method:
- according to manual
- 20µl of re-suspended cell-suspension were plated on a LB-Amp-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones
modified PCR to generate biobrick-ready genes from geneart-nanobody construct
Investigators: Sascha
Materials:
- Templates: digested geneart-nanobody construct
- Phusion-polymerase
- 10x Phusion buffer HF
- DMSO
- dNTPs (10mM)
- Primer 2.1 and 2.2: TEV-LoxP-TMD-mcherry-LoxP
- Primer 4.1 and 4.2(=1.2): Fc
- Primer5.1 and 5.2: TMD-mcherry
- Thermocycler
Methods: 50µl mix for every biobrick-ready gene, PCR-mix applies for all genes
| reagent | volume [µL]' |
| 10x Phusion HF-buffer | 10 |
| dNTPs | 1 |
| Primer5.1 : TMD-mcherry ; Primer 2.1: TEV-LoxP-TMD-mcherry-LoxP ; 4.1: Fc | 2,5 |
| Primer 5.2: TMD-mcherry ; Primer 2.2: TEV-LoxP-TMD-mcherry-LoxP ; Primer 4.2(=1.2): Fc | 2,5 |
| digested geneart-nanobody construct (10ng/µl) | 2 |
| Phusion Polymerase | 0,5 |
| water | 32,5 |
Program for gene2,4
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 15 |
| annealing | gradient: 55°C, 60°C, 70°C | 10 | 15 |
| elongation | 72 | 20 | 15 |
| denaturation | 98 | 8 | 15 |
| annealing/elongation | 72 | 20 | 15 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: analytical gelelectrophoresis of amplified gene 2,4
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
- amplified DNA
Method:
- 1% agarosegel, 90ml
- 2µl of each PCR-mix, 1µl FD Green, 8µl water
- 115V,80min
Results:
- only gene5 (~800bp) show DNA at desired bp-size
Further Tasks:
- furhter amplification of genes 2, 4
modified PCR to generate biobrick-ready genes (2 and 4) from geneart-nanobody construct
Investigators: Sascha
Materials:
- Templates: digested geneart-nanobody construct
- Phusion-polymerase
- 10x Phusion buffer HF
- DMSO
- dNTPs (10mM)
- Primer 2.1 and 2.2: TEV-LoxP-TMD-mcherry-LoxP
- Primer 4.1 and 4.2(=1.2): Fc
- Thermocycler
Methods: 50µl mix for every biobrick-ready gene, PCR-mix applies for all genes
| reagent | volume [µL]' |
| 10x Phusion HF-buffer | 10 |
| dNTPs | 1 |
| Primer 2.1: TEV-LoxP-TMD-mcherry-LoxP ; 4.1: Fc | 2,5 |
| Primer 2.2: TEV-LoxP-TMD-mcherry-LoxP ; Primer 4.2(=1.2): Fc | 2,5 |
| digested geneart-nanobody construct (10ng/µl)td> | 2 |
| Phusion Polymerase | 0,5 |
| water | 32,5 |
Program for gene2,4
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 15 |
| annealing | gradient: 55°C, 60°C, 70°C | 10 | 15 |
| elongation | 72 | 20 | 15 |
| denaturation | 98 | 8 | 15 |
| annealing/elongation | 72 | 20 | 15 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
2012-09-12
preparative digestion of scFV construct with AgeI and XbaI
Investigator: Maria
Aim: digestion of scFv construct with AgeI and XbaI to generate sticky ends for ligation into pSB1C3
Materials:
- Fast Digest AgeI
- Fast Digest XbaI
- 10x FD Green Buffer
- scFv construct
- sterile water
Method:
2x
- 20µl scFv construct
- 2µl NheI
- 2µl ApaI
- 3µl 10x FD Green Buffer
- 5µl sterile water
- digestion for 2,5h at 37°C
Further Tasks:
- gelelectrophoresis
- gelextraction
preparative gelelectrophoresis of digested scFv-TEV-TMD-EYFP
Investigator: Maria
Aim: preparative gelelectrophoresis to separate digested scFv-TEV-TMD-EYFP construct from cutted overhangs in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
Method:
- 2x 30µl digested scFv construct
- 1% agarose gel, 100ml
- 100V
Results:
- digested scFv construct
Further Tasks:
- gelextraction
- ligation
gelextracton of digested scFv-TEV-TMD-EYFP construct out of 1% agarosegel
Investigators:Maria
Aim: gelextraction and preparation of cleaned scFv-TEV-TMD-EYFP construct
Materials:
- Gel-Clean-Up Kit
Method:
- according to manual
Results:
- 8,3 ng/µl
Further tasks:
- ligation into pSB1C3
analytical gelelectrophoresis of scFv-construct with modified PCR mix
Investigator: Maria
Aim: analytical gelelectrophoresis to check PCr result
Materials:
- agarose
- 1xTAE-buffer
- 10xFD FDGreen
Method:
- 3µl 10x FD Green, 15µl of each PCR product
- 1% agarose gel, 100ml
- 120V
Results:
- no precise bands
assembly PCR of mutated scFv without PstI out of part 1 (primer 1 and 2) and part 2 (primer 3 and 4) and mutated scF without PstI (primer 1 and 4)
Investigator: Maria
AIM: assembly of scFV without PstI restriction sites out of part 1 and 2 + multiplication of mutated scFv construct without PstI sites
Materials:
- Template mutated scFv part 1 and 2, mutated scFv construct
- Phusion-polymerase
- 10x Phusion buffer GC
- dNTPs (10mM)
- forward primer: small-I-fw-prefix25 (Tm=72°C)
- reverse primer: small-6-rw-suffix-25 (Tm= 72°C)
- DSMSO
- Thermocycler
Methods:
Mastermix for multiplication of mutated scFv
| reagent | volume [µL] |
| 10x Phusion GC buffer | 10 |
| dNTPs | 1 |
| Primer (Forward) | 2,5 |
| Primer (Reverse) | 2,5 |
| template 1 | 1 |
| Phusion Polymerase | 0,5 |
| water | 32,5 |
Mastermix for triplex batch assembly part 1 and 2
| reagent | volume [µL] |
| 10x Phusion GC buffer | 30 |
| dNTPs | 3 |
| Primer (Forward) | 7,5 |
| Primer (Reverse) | 7,5 |
| template 1 | 1,75 (5,7 ng/µl) |
| template 2 | 1,36 (3,4 ng/µl) |
| DMSO | 4,5 |
| Phusion Polymerase | 1,5 |
| water | 86,67 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | 71 | 10 | 5 |
| annealing + elongation | 72 | 30 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing | 72 | 35 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Further tasks:
- analytical gelelectrophoresis
picking clones of cre recombinase and BBa_404316, inoculation of o.n.-culture
Investigator: Maria
Materials:
- 15 ml inoculation tubes
- LB-Medium
- Ampicillin
- bunsen burner
Method:
- 1x o.n.-culture of cre recombinase
- 1x o.n.-culture of BBa-404316
- each o.n.-culture 5ml LB-Medium + 5µl Ampicillin
- incubation o.n. at 37°C
Further tasks:
- miniprep of o.n.-cultures
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO cells w/ zeocin and w/o zeocin
- seeding CHO cells in Ibidi Dish for transfection and Virus infection experiment
- dilution of stably transfected CHO Clone 29 into 75cm² flasks (+550µg/ml Hygromycin)(according to protocol)
- co-transfection of Nanobody construct clone 29 and modified AID with eGFP (according to protocol)
Topic: analytical gel electrophoresis of amplified genes 2 and 4 and preparative gel electrophoresis of digested geneart-nanobody construct
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
- amplified DNA
Method:
- 1% agarosegel, 100ml
- 2µl of each PCR-mix, 1µl FD Green, 8µl water
- 110V,80min
Results:
Further Tasks:
- amplification of gene 1
PCR to generate biobrick-ready gene 1 from geneart-nanobody construct
Investigators: Sascha
Materials:
- Templates: geneart-nanobody construct
- Phusion-polymerase
- 10x Phusion buffer HF
- DMSO
- dNTPs (10mM)
- Primer 1.1 and 1.2: Signal peptide-Nanobody-Fc
- Thermocycler
Methods: 4x 50µl PCR-mix for gene1
| reagent | volume [µL]' |
| 10x Phusion HF-buffer | 40 |
| dNTPs | 4 |
| Primer 1.1: Signal peptide-Nanobody-Fc | 10 |
| Primer 1.2: Signal peptide-Nanobody-Fc | 10 |
| geneart-nanobody construct (25ng/µl) | 1 |
| Phusion Polymerase | 2 |
| water | 133 |
- PCR-Mastermix of gene1 was split into 4x 50µl mixes
Program for gene1
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | gradient: 55°C, 60°C, 70°C | 10 | 15 |
| elongation | 72 | 20 | 15 |
| denaturation | 98 | 8 | 15 |
| annealing/elongation | 72 | 20 | 15 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: analytical gel electrophoresis of amplified genes 2 and 4 and preparative gel electrophoresis of digested geneart-nanobody construct
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
- amplified DNA
- digested geneart-nanobody construct (~2,2µg)
Method:
- 1% agarosegel, 100ml
- 2µl of each PCR-mix, 1µl FD Green, 8µl water
- 100V,95min
Results:
Further Tasks:
- PCR-clean up of genes 1,4,5
- digestion of genes 1,4,5 and psB1C3 with XbaI and AgeI
2012-09-13
Glycerol stock and Mini Prep of cre recombinase, BBa_404316 and YFP
Investigator: Maria
Materials: overnight cultures, Mini prep kit
Methods: according to manual
Results:
Cre recombinase : 192,8 ng/µl
BBa_404316: 242,5 ng/µl<br<
YFP: 130,6 ng/µl
Glycerol stocks of cre recombinase and BBa_404316
Further tasks: preparative digestion of BBa_404316 and 50ml culture of cre recombinase for endotoxin free preparation
analytical gelelectrophoresis of assembly PCR result scFv only (primer 7 and 8)
Investigator: Maria
Aim: analytical gelelectrophoresis to check PCR result
Materials:
- agarose
- 1xTAE-buffer
- 10xFD FDGreen
Method:
- 1µl 10x FD Green, 1µl of each PCR product, add sterile water to 10 µl
- 1% agarose gel, 100ml
- 120V
Results:
- precise bands
Further Tasks:
- preparative gelelectrophoresis
preparative gelelectrophoresis of PCR result for scFV only (primer 7 and 8)
Investigator: Maria
Aim: preparative gelelectrophoresis of assembled scFv only in 1% agarose gel
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
Method:
- 2x 30µl of scFv only
- 1% agarose gel, 100ml
- 100V
Further Tasks:
- gelextraction
- ligation
gelextracton of PCR product scFv only out of 1% agarosegel
Investigator: Maria
Aim: gelextraction and preparation of cleaned scFv only construct
Materials:
- Gel-Clean-Up Kit
Method:
- according to manual
Results:
- 1: 86,1 ng/µl
- 2: 108,2 ng/µl
Further Tasks:
- digestion with AgeI and XbaI
- ligation into pSB1C3
preparative digestion of scFV only and BBa_404316 with AgeI and XbaI
Investigator: Maria
Aim: digestion of scFv only and BBa_404316 (pSB1C3 with RCF25 standard) with AgeI and XbaI
Materials:
- Fast Digest AgeI
- Fast Digest XbaI
- 10x FD Green Buffer
- scFv only
- BBa_404316
- steril water
Method:
2x
- 8,25µl BBa_404316
- 1µl AgeI
- 1µl XbaI
- 3µl 10x FD Green Buffer
- 16,75 µl sterile water
1x
- 13,8µl scFv only
- 1µl AgeI
- 1µl XbaI
- 3µl 10x FD Green Buffer
- 11,2µl sterile water
- digestion for 2,5h at 37°C
- heat inactivation at 65°C for 5 min
Further Tasks:
- PCR clean-up of digested scFV only
- dephosphorylation of pSB1C3
- gelelectrophoresis
- gelextraction
PCR-Clean-Up of digested scFv only construct
Investigator:Maria
Aim: purifying scFv only construct
Materials:
- PCR-Clean-Up Kit
Method:
- according to manual
Results:
- concentration of digested scFv only 66,8 ng/µl
dephosphorylating the digested pSB1C3 vector
Investigator:Maria
Aim:dephosphorylating the digested pSB1C3 vector to prevent re-ligation
Materials:
- Antarctic Phosphatase
- 10x Antarctic Phosphatase Reaction Buffer
- digested pSB1C3
Method:
- 3,3µl 10x Antarctic Phosphatase Reaction Buffer
- 1µl Antarctic Phosphatase
- incubation for 30min at 37°C to dephosphorylate 5´-ends of pSB1C3
- heat inactivation of Phosphatase for 5min at 65°C
- store at -20°C
Further Tasks:
- preparative gelelectrophoresis
- gelextraction
preparative gelelectrophoresis of digested and dephosphorylated pSB1C3
Investigators:Maria
Aim: preparative gelelectrophoresis of digested and dephosphorylated pSB1C3 in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
Method:
- 2x 30µl digested pSB1C3
- 1% agarose gel, 100ml
- 100V
Results:
- cleaned and digested pSB1C3 and scFv only construct
Further Tasks:
- gelextraction
- ligation
gelextracton of digested and dephosphorylated pSB1C3 out of 1% agarose gel
Investigator: Maria
Aim: gelextraction and preparation of digested and dephosphorylated pSB1C3
Materials:
- Gel-Clean-Up Kit
Method:
- according to manual
Results:
- I: 49,7 ng/µl, II: 53,3ng/µl
Further Tasks:
- ligation with scFv only and scFv construct
ligation of digested scFv-only and scFV construct and dephosporylated pSB1C3
Investigator: Maria
Aim: ligation of digested scFv-only and scFv construct with pSB1C3
Materials:
- ligation calculator: http://www.insilico.uni-duesseldorf.de/Lig_Input.html
- T4 DNA ligae
- ligase buffer
- digested scFv-only and scFv construct
- digested and dephosporylated pSB1C3
Method: ligation-ratio--> 1:3
I scFv only:
- 1µl T4 DNA ligase
- 2,5µl T4 DNA-ligase buffer
- 1,24µl digested scFv only (22,26ng/µl)
- 1µl digested pSB1C3 (26,65ng/µl)
- 14,26µl sterile water
II scFv construct:
- 1µl T4 DNA ligase
- 2,5µl T4 DNA-ligase buffer
- 4,81µl digested scFv construct (13,1ng/µl)
- 1µl digested pSB1C3 (26,65ng/µl)
- 10,69µl sterile water
- incubation for 1h at RT
Further Tasks:
- transformation of ligation mix into XL1-blue competent E. coli cells
Transformation of ligated pSB1C3-scFv-only and pSB1C3-scFv-construct into new XL1-blue competent E. coli cells
Investigator: Maria
Materials:
- Bunsen Burner, Agar Plate with Ampicillin, 37 °C thermo mixer, centrifuge,
- 10 µl of pSB1C3-scFv-only and pSB1C3-scFv-construct
- icebox
- new competent E. coli cells (XL 1)
Method:
- according to manual
- 20µl of resuspended cell-suspension were plated on a LB-Cm-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones
cell culture
Investigator: Kerstin
Topic: cell-culture
- passaging of CHO cells w/ zeocin and w/o zeocin
- seeding HT1080 in Ibidi Dish for Virus infection experiment
- transfection of CHO cells in Ibidi Dish with Nanobody construct (according to protocol)
- passaging of HEK AAV 293
- registration at FACS core facility at Deutsches Rheumaforschungszentrum
- planning of further experiments
PCR-Clean up genes biobrick-ready genes 1,4,5
Investigator: Sascha
Materials:
- NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel)
Methods:
- according to CleanUp-protocol
Gene 1 (Signal peptide-Nanobody-Fc) = 112,4ng/µl
Gene 4 (Fc): = 17,1ng/µl
Gene 5 (TMD-mcherry) = 193ng/µl
Further Taks:
- preparative digestion of genes 1,4,5 with XbaI and AgeI
preparative digestion of genes 1,4 and 5 with XbaI and AgeI
Investigator: Sascha
Materials:
- Fast Digest XbaI
- Fast Digest AgeI
- 10x FD Green Buffer
- genes 1,4,5
- sterile water
Method:
2x
- 15µl of gene1 (112,4ng/µl)
- 10µl og gene 5 (193ng/µl)
- 25µl of gene4 (17,1ng/µl)
- 2µl NheI
- 2µl ApaI
- 4µl 10x FD Green Buffer
- sterile water ad 40µl
- digestion for 2h at 37°C
>Results:
Further Tasks:
- PCR-clean up of digested genes 1,4,5
preparative gel electrophoresis followed by gel extraction of digested genes 1,4,5
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
- amplified DNA
- digested genes 1,4,5
Method:
- 1% agarosegel, 100ml
- whole digestion-mix of each gene
- 100V,95min
- desired DNA bands for gene1: approximately 880bp, gene 4: 700bp, gene5: 800
Materials:
- NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel)
Methods:
- according to gel extraction-protocol
Gene 1 (Signal peptide-Nanobody-Fc) = 18,3ng/µl, R=1,75
Gene 4 (Fc): = 10,4ng/µl, R= 1,72
Gene 5 (TMD-mcherry) = 68,1ng/µl, R= 1,82
Further Taks:
- ligation of digested genes 1,4,5 into digested psB1C3 (X+A)
modified PCR to generate biobrick-ready genes (2 and 3) from geneart-nanobody construct
Investigators: Sascha
Materials:
- Templates: digested geneart-nanobody construct
- Phusion-polymerase
- 10x Phusion buffer HF
- dNTPs (10mM)
- Primer 2.1 and 2.2: TEV-LoxP-TMD-mcherry-LoxP
- Primer 3.1 and 3.2: Nanobody
- Thermocycler
Methods: 4x 50µl PCR-mastermix for gene2 and 3, PCR-mix applies for all genes
| reagent | volume [µL]' |
| 10x Phusion HF-buffer | 10 |
| dNTPs | 1 |
| Primer 2.1: TEV-LoxP-TMD-mcherry-LoxP ; Primer 3.1: Nanobody | 2,5 |
| Primer 2.2: TEV-LoxP-TMD-mcherry-LoxP ; Primer 3.2: Nanobody | 2,5 |
| digested geneart-nanobody construct (10ng/µl)td> | 2 |
| Phusion Polymerase | 0,5 |
| water | 32,5 |
Program for gene2
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 15 |
| annealing | gradient: 55°C, 60°C, 70°C | 10 | 15 |
| elongation | 72 | 20 | 15 |
| denaturation | 98 | 8 | 15 |
| annealing/elongation | 72 | 20 | 15 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Program for gene3
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 5 |
| annealing | gradient: 55°C, 60°C, 70°C | 10 | 5 |
| elongation | 72 | 6 | 5 |
| denaturation | 98 | 8 | 25 |
| annealing/elongation | 72 | 8 | 25 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
2012-09-14
Transformation of ligated pSB1C3-scFv-only and pSB1C3-scFv-construct into new XL1-blue competent E. coli cells
Investigator: Maria
Materials:
- Bunsen Burner, Agar Plate with Ampicillin, 37 °C thermo mixer, centrifuge,
- 10 µl of pSB1C3-scFv-only and pSB1C3-scFv-construct
- icebox
- new competent E. coli cells (XL 1)
Method:
- according to manual
- 20µl of resuspended cell-suspension were plated on a LB-CM-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO cells w/ zeocin and w/o zeocin
- dilution of stably transfected cells (scFv construct clone 4, originally from transfected Well 3) to 1-2 cells per 150µl and seeding in 3x 96-Well plates
- passaging of HT1080
- infection of CHO cells in Ibidi Dish with Virus (YFP on surface and CFP as GOI)(according to protocol)
- continuation of G418 selection experiment
- planning of further experiments
Topic: analytical gel electrophoresis of amplified genes 2 and 3
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
- amplified DNA
Method:
- 1% agarosegel, 100ml
- 2µl of each PCR-mix, 1µl FD Green, 8µl water
- 110V,85min
Results:
- gene 3 shows sufficient bp-size (~350bp)
Further Tasks:
- further PCR of gene2
Topic: ligation of digested dephosporylated pSB1C3 (X+A) and digested genes 1,4,5 (X+A)
Investigator: Sascha
Materials:
- ligation calculator: http://www.insilico.uni-duesseldorf.de/Lig_Input.html
- T4 DNA ligae
- ligase buffer
- digested genes 1,4,5
- digested and dephosporylated pSB1C3
Method: ligation-ratio--> 1:3
1st ligation mix-gene1:
- 1µl T4 DNA ligase
- 2µl T4 DNA-ligase buffer
- 20 ng dig. psB1C3
- 32,3 ng insert (gene1)
- sterile water ad 20µl
2nd ligation mix-gene4:
- 1µl T4 DNA ligase
- 2µl T4 DNA-ligase buffer
- 20 ng dig. psB1C3
- 32,3 ng insert (gene4)
- sterile water ad 20µl
3rd ligation mix-gene1:
- 1µl T4 DNA ligase
- 2µl T4 DNA-ligase buffer
- 20 ng dig. psB1C3
- 22,9 ng insert (gene5)
- sterile water ad 20µl
- incubation for 40min at RT
Further Tasks:
- transformation of ligation mix into XL1-blue competent E. coli cells
Transformation of ligated pSB1C3-gene 1, 4, 5 into new XL1-blue competent E. coli cells
Investigator: Sascha
Materials:
- Bunsen Burner, Agar Plate with Ampicillin, 37 °C thermo mixer, centrifuge,
- 15 µl of each ligation mix
- icebox
- new competent E. coli cells (XL 1)
Method:
- according to manual
- 20µl of resuspended cell-suspension were plated on a LB-Cm-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones
modified PCR to generate biobrick-ready genes 2 from geneart-nanobody construct
Investigators: Sascha
Materials:
- Templates: digested geneart-nanobody construct
- Phusion-polymerase
- 10x Phusion buffer HF
- dNTPs (10mM)
- Primer 2.1 and 2.2: TEV-LoxP-TMD-mcherry-LoxP
- Thermocycler
Methods: 4x 50µl PCR-mastermix for gene2 , PCR-mix applies for all genes
| reagent | volume [µL] |
| 10x Phusion HF-buffer | 10 |
| dNTPs | 1 |
| Primer 2.1: TEV-LoxP-TMD-mcherry-LoxP | 2,5 |
| Primer 2.2: TEV-LoxP-TMD-mcherry-LoxP | 2,5 |
| digested geneart-nanobody construct (10ng/µl) | 2 |
| Phusion Polymerase | 0,5 |
| water | 32,5 |
Program for gene2
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 15 |
| annealing | gradient: 55°C, 60°C, 70°C | 10 | 15 |
| elongation | 72 | 20 | 15 |
| denaturation | 98 | 8 | 15 |
| annealing/elongation | 72 | 20 | 15 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
2012-09-15
picking clones of ligated scFv-only-pSB1C3 and scFv-construct-pSB13C
Investigator: Kathi
Materials:
- 11x 15 ml inoculation tubes
- LB-Medium
- Chloramphenicol
- bunsen burner
Method:
- 5x o.n.-culture of scFv-only-pSB1C3
- 5x o.n.-culture of scFv-construct-pSB1C3
- each o.n.-culture 5ml LB-Medium + 5µl Chloramphenicol
- incubation o.n. at 37°C
Further Tasks:
- miniprep of o.n.-cultures
picking clones of ligated pSB1C3-genes1,4 and 5
Investigator: Sascha
Materials:
- 3x5 ml inoculation tubes
- LB-Medium
- Chloramphenicol
- bunsen burner
Method:
- 5x o.n.-culture of pSB1C3-gene1(Signal peptide-Nanobody-Fc)
- 5x o.n.-culture of pSB1C3-gene4(Fc)
- 5x o.n. culture of psB1C3-gene5(TMD-mcherry)
- each o.n.-culture 5ml LB-Medium + 5µl Chloramphenicol
- incubation o.n. at 37°C
Further Tasks:
- miniprep of o.n.-cultures
Topic: analytical gel electrophoresis of amplified genes 2
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
- amplified DNA
Method:
- 1% agarosegel, 100ml
- 2µl of each PCR-mix, 1µl FD Green, 8µl water
- 110V,85min
Results:
Further Tasks:
- PCR-Clean up
PCR-clean up of digested genes 2 and 3
Investigator: Sascha
Materials:
- NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel)
Methods:
- according to CleanUp-protocol
Gene 2 (Signal peptide-Nanobody-Fc) = --ng/µl, R=-- Gene 3 (Fc): = --ng/µl, R= --
Further Taks:
- preparative digestion of genes 2 and 3 with XbaI and AgeI
preparative digestion of genes 2 and 3 with XbaI and AgeI
Investigator: Sascha
Materials:
- Fast Digest XbaI
- Fast Digest AgeI
- 10x FD Green Buffer
- genes 1,4,5
- sterile water
Method:
2x
- --µl of gene1 (--ng/µl)
- --µl og gene 5 (---ng/µl)
- 25µl of gene4 (---ng/µl)
- 2µl NheI
- 2µl ApaI
- 4µl 10x FD Green Buffer
- sterile water ad 40µl
- digestion for 2h at 37°C
Results:
Further Tasks:
- preparative gel electrophoresis and gel extraction of digested gene2 and 3
preparative gel electrophoresis followd by gel extraction of digested genes 2 and 3
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
- amplified DNA
- digested genes 2 and 3
Method:
- 1% agarosegel, 100ml
- whole digestion-mix of each gene
- 100V,95min
Results:
Materials:
- NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel)
Methods:
- according to gel extraction-protocol
Gene 2 (TEV-LoxP-TMD-mcherry-LoxP ) = 18,3ng/µl, R=1,75
Gene 3 (Nanobody): = 10,4ng/µl, R= 1,72
Further Taks:
- ligation of digested genes 2 and 3 into digested psB1C3 (X+A)
2012-09-16
Glycerol stock and Mini Prep of scFv-only-pSB1C3 and scFv-construct-pSB1C3 clones
Investigator: Maria
Materials: overnight cultures scFv-only-pSB1C3 and scFv-construct-pSB1C3 clones, Mini prep kit
Methods: according to manual
Results:
- scFv-only-pSB1C3
Clon 1: 288,3ng/µl
Clon 2: 309,8ng/µl<br<
Clon 3: 408,2ng/µl
Clon 4: 100,7ng/µl
Clon 5: 363,8ng/µl
- scFv-construct-pSB1C3
Clon 1: 398,8ng/µl
Clon 2: 439,3ng/µl
Clon 3: 337,4ng/µl
Clon 4: 80ng/µl
Clon 5: 125,9ng/µl
Glycerole stocks of all clones
Further Tasks: analytical digestion, analytical gelelectrophoresis
analytical digestion of ligated scFv-only-pSB1C3 and scFv-construct-pSB1C3 with PstI and XbaI
Investigator: Maria
Materials:
- all 10 prepared scFv-only-pSB1C3 and scFv-construct-pSB1C3 clones
- pSB1C3 vector control
- FastDigest PstI and XbaI
- 10x FD Green Buffer
- sterile water
Method:
- 10µl mix: 1µl PstI, 1µl 10x FD Green Buffer, 1µl of each clone (1:2, approximately 200ng DNA), 7µl sterile water
- 11µl mix: 1µl XbaI, 1µl 10x FD Green Buffer, 1µl of each clone (1:2, approximately 200ng DNA), 7µl sterile water
- incubation at 37°C for 45 min
Further Tasks:
- analytical gelelectrophoresis
Topic: gelelectrophoresis of analytical digested scFv-only-pSB1C3 and scFv-construct-pSB1C3 clones
Investigator: Maria
Aim: checking plasmid-size after ligation in 1% agarose gel
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarose gel, 100ml
- 120 V
Results:
- ligation partly successful
Further Tasks:
- preparation for sequencing
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging HEK AAV 293
- passaging HT1080
- change of medium in 75cm² flasks with stably transfected CHO clone 29 and clone 4 (+550µg/ml Hygromycin)
- capturing images of virus-infection in Ibidi Dish 8see Results)
- continuation of G418 selection experiment
- seeding CHO in Ibidi Dishes for transfection and virus infection experiment
ligation of digested dephosporylated pSB1C3 (X+A) and digested genes 2 and 3 (X+A)
Investigator: Sascha
Materials:
- ligation calculator: http://www.insilico.uni-duesseldorf.de/Lig_Input.html
- T4 DNA ligase
- ligase buffer
- digested genes 2 and 3
- digested and dephosporylated pSB1C3
Method: ligation-ratio--> 1:3
1st ligation mix-gene2:
- 1µl T4 DNA ligase
- 2µl T4 DNA-ligase buffer
- 20 ng dig. psB1C3
- 27 ng insert (gene1)
- sterile water ad 20µl
2nd ligation mix-gene3:
- 1µl T4 DNA ligase
- 2µl T4 DNA-ligase buffer
- 20 ng dig. psB1C3
- 10,5 ng insert (gene3)
- sterile water ad 20µl
- incubation for 40min at RT
Further Tasks:
- transformation of ligation mix into XL1-blue competent E. coli cells
Glycerol stock and Mini Prep of psB1C3-gene1,4 and 5 constructs
Investigator: Sascha
Materials: overnight cultures of psB1C3-gene1,4 and 5 constructs, Mini prep kit
Methods: according to manual
Results:
- psB1C3-gene1
Clon 1: ng/µl
Clon 2: ng/µl
Clon 3: ng/µl
Clon 4: ng/µl
Clon 5: ng/µl
- psB1C3-gene4
Clon 1: ng/µl
Clon 2: ng/µl
Clon 3: ng/µl
Clon 4: ng/µl
Clon 5: ng/µl
- psB1C3-gene5
Clon 1: ng/µl
Clon 2: ng/µl
Clon 3: ng/µl
Clon 4: ng/µl
Clon 5: ng/µl
Glycerole stocks of all clones
Further Tasks:
- test digestion, analytical gelelectrophoresis
test digestion of psB1C3-gene1,4 and 5 constructs XbaI and AgeI
Investigator: Sascha
Materials:
- Fast Digest XbaI
- Fast Digest AgeI
- 10x FD Green Buffer
- psB1C3-gene1,4 and 5 constructs
- sterile water
Method:
Mastermix for test digestion: for 16 reactions; subsequently specified for one reaction
- ~ 100ng DNA
- 1µl NheI
- 1µl ApaI
- 1µl 10x FD Green Buffer
- sterile water ad 10µl
- digestion for 30min at 37°C
Results:
Further Tasks:
- analytical gel electrophoresis
Topic: analytical gel electrophoresis of psB1C3-gene1,4 and 5 constructs
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
Method:
- 1% agarosegel, 100ml
- 10µl of each digestion mix
- 110V,85min
Results:
- all genes show satisfaying bp-size
- gene 1.2, gene1.5, gene4.2 and gene5.2 picked up for sequencing
Further Tasks:
- sequencing
planing immunostaining to detect expressed scFv on surface of CHO cells
Investigator: Kerstin, Sascha
Materials:
- pubmed
- online data bases
2012-09-17
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO cells w/ zeocin
- change of medium in 75cm² flasks with stably transfected CHO clone 29 (+550µg/ml Hygromycin)
- capturing images of virus-infection in Ibidi Dish (see Results)
- seeding cells for co-transfection with Nanobody construct and Cre Recombinase
choosing clones and preparing ligated psB1C3-genes1,4,5, scFv-only and scFv-construct for ordering sequencing by GATC
Investigators: Sascha
Materials:
- dilute DNA according to GATC requirements
- Geneious
- Using psB1C3 primer fw/rv
Further Tasks:
- analysis of sequencing results
preparative digestion of geneart-nanobody-construct with XbaI and AgeI
Investigator: Sascha
Materials:
- Fast Digest XbaI
- Fast Digest AgeI
- 10x FD Green Buffer
- geneart-nanobody-construct
- sterile water
Method:
2x digestoin mix
- ~2,3µg geneart-nanobody-construct
- 2µl NheI
- 2µl ApaI
- 4µl 10x FD Green Buffer
- sterile water ad 40µl
- digestion for 2h at 37°C
Results:
Further Tasks:
- preparative gel electrophoresis and gel extraction of digested geneart-nanobody-construct
preparative gel electrophoresis followed by gel extraction of digested geneart-nanobody-construct
Investigator: Basia/ Sascha
Materials:
- agarose
- 1xTAE-buffer
- amplified DNA
- geneart-nanobody-construct
Method:
- 1% agarosegel, 100ml
- whole digestion-mix of each gene
- 100V,95min
Results:
Materials:
- NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel)
Methods:
- according to gel extraction-protocol
geneart-nanobody-construct = ng/µl, R=
Further Tasks:
- ligation of digested geneart-nanobody-construct into digested psB1C3 (X+A)
ligation of digested dephosporylated pSB1C3 (X+A) and digested geneart-nanobody-construct (X+A)
Investigator: Basia
Materials:
- ligation calculator: http://www.insilico.uni-duesseldorf.de/Lig_Input.html
- T4 DNA ligase
- ligase buffer
- digested geneart-nanobody-construct
- digested and dephosporylated pSB1C3
Method: ligation-ratio--> 1:3
1st ligation mix-gene2:
- 1µl T4 DNA ligase
- 2µl T4 DNA-ligase buffer
- 20 ng dig. psB1C3
- ng insert (geneart-nanobody-construct )
- sterile water ad 20µl
- incubation for 40min at RT
Further Tasks:
- transformation of ligation mix into XL1-blue competent E. coli cells
Transformation of ligated pSB1C3-geneart-nanobody-construct into new XL1-blue competent E. coli cells
Investigator: Basia
Materials:
- Bunsen Burner, Agar Plate with Ampicillin, 37 °C thermo mixer, centrifuge,
- 15 µl of each ligation mix
- icebox
- new competent E. coli cells (XL 1)
Method:
- according to manual
- 20µl of resuspended cell-suspension were plated on a LB-Cm-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones
picking clones of ligated pSB1C3-genes2 and 3
Investigator: Sascha
Materials:
- 2x 5 ml inoculation tubes
- LB-Medium
- Chloramphenicol
- bunsen burner
Method:
- 5x o.n.-culture of pSB1C3-gene2(TEV-LoxP-TMD-mcherry-Lox)
- 5x o.n.-culture of pSB1C3-gene3(Nanobody)
- each o.n.-culture 5ml LB-Medium + 5µl Chloramphenicol
- incubation o.n. at 37°C
Further Tasks:
- miniprep of o.n.-cultures
planing immunostaining to detect expressed scFv on surface of CHO cells
Investigator: Kerstin, Sascha
Materials:
- pubmed
- online data bases
2012-09-18
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO cells w/ zeocin
- passaging of HEK AAV 293
- passaging of HT1080
- continuation of G418 selection experiment
- seeding CHO cells for transfection and FACS
- seeding cells for co-transfection with Nanobody construct and Cre Recombinase
Glycerol stock and Mini Prep of psB1C3-gene 2,3 constructs
Investigator: Sascha
Materials: overnight cultures of psB1C3-gene2,3 constructs, Mini prep kit
Methods: according to manual
Results:
- psB1C3-gene2
Clon 1: ng/µl
Clon 2: ng/µl
Clon 3: ng/µl
Clon 4: ng/µl
Clon 5: ng/µl
- psB1C3-gene3
Clon 1: ng/µl
Clon 2: ng/µl
Clon 3: ng/µl
Clon 4: ng/µl
Clon 5: ng/µl
Glycerole stocks of all clones
Further Tasks:
- test digestion, analytical gelelectrophoresis
test digestion of psB1C3-gene2 and 3 constructs XbaI and AgeI
Investigator: Sascha
Materials:
- Fast Digest XbaI
- Fast Digest AgeI
- 10x FD Green Buffer
- psB1C3-gene1,4 and 5 constructs
- sterile water
Method:
Mastermix for test digestion: for 11 reactions; subsequently specified for one reaction
- ~ 100ng DNA
- 1µl NheI
- 1µl ApaI
- 1µl 10x FD Green Buffer
- sterile water ad 10µl
- digestion for 30min at 37°C
>Results:
Further Tasks:
- analytical gel electrophoresis
Topic: analytical gel electrophoresis of psB1C3-gene2 and 3constructs
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
Method:
- 1% agarosegel, 90ml
- 10µl of each digestion mix
- 115V,80min
- all genes show satisfaying bp-size
- gene 2.5 and gene3.3 picked up for sequencing
Further Tasks:
- sequencing
choosing clones and preparing ligated psB1C3-genes2, 3 construct for ordering sequencing by GATC
Investigators: Sascha
Materials:
- dilute DNA according to GATC requirements
- Geneious
- Using psB1C3 primer fw/rv
Further Tasks:
- analysis of sequencing result
picking clones of ligated pSB1C3-geneart-nanobody-construct
Investigator: Sascha
Materials:
- 1x 5 ml inoculation tubes
- LB-Medium
- Chloramphenicol
- bunsen burner
Method:
- 5x o.n.-culture of pSB1C3-geneart-nanobody construct
- each o.n.-culture 5ml LB-Medium + 5µl Chloramphenicol
- incubation o.n. at 37°C
Further Tasks:
- miniprep of o.n.-cultures
planing immunostaining to detect expressed scFv on surface of CHO cells
Investigator: Kerstin, Sascha
Materials:
- pubmed
- online data bases
Results:
- Pacific Blue goat anti-mouse IgG
2012-09-19
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO cells w/ zeocin and w/o zeocin
- change of medium in 75cm² flasks with stably transfected CHO clone 4 (+550µg/ml Hygromycin)
- co-transfection of CHO cells with nanobody construct (clone 29) and Cre Recombinase in 2x 25cm² culture flasks
- HEK 293 (20000 cells/well) and HeLa (10000 cells/well) were seeded in Ibidi Dish for transfection
- first attempt at live-cell immunfluorescence: incubation with primary antibody (anti-flag-tag antibody) in serum-free medium for 1h on shaker, after careful washing incubation with secondary antibody (Pacific Blue Alexa Fluor®) for 1h, capturing of images was done with the fluorescence microscope
looking for new transmembrane domains and signal peptides
Investigator: Sascha
Materials:
- pubmed
- online data bases
Glycerol stock and Mini Prep of psB1C3-nanobody construct
Investigator: Sascha
Materials: overnight cultures of psB1C3-nanobody construct, Mini prep kit
Methods: according to manual
Results:
- psB1C3-nanobody construct
Clon 1: ng/µl
Clon 2: ng/µl<br<
Clon 3: ng/µl
Clon 4: ng/µl
Clon 5: ng/µl
Glycerole stocks of all clones
Further Tasks:
- test digestion, analytical gelelectrophoresis
test digestion of psB1C3-nanobody construct with XhoI
Investigator: Sascha
Materials:
- Fast Digest XhoI
- 10x FD Green Buffer
- psB1C3-nanobody
- sterile water
Method:
Mastermix for test digestion: for 6 reactions; subsequently specified for one reaction
- ~ 100ng DNA
- 1µl XhoII
- 1µl 10x FD Green Buffer
- sterile water ad 10µl
- digestion for 30min at 37°C
Results:
Further Tasks:
- analytical gel electrophoresis
Topic: analytical gel electrophoresis of psB1C3-nanobody construct
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
Method:
- 1% agarosegel, 90ml
- 10µl of each digestion mix
- 115V,80min
- selecting clone 1 for sequencing
Further Tasks:
- sequencing
choosing clones and preparing ligated psB1C3-nanobody construct for ordering sequencing by GATC
Investigators: Sascha
Materials:
- dilute DNA according to GATC requirements
- Geneious
- Using psB1C3 primer fw/rv
Further Tasks:
- analysis of sequencing result
planing of detecting nanobody and scFv in transfected CHO cells
Investigator: Kerstin, Sascha
Materials:
- pubmed
- online data bases
2012-09-20
cell culture
Investigator:Kerstin
Topic: cell-culture
- capturing images of stably transfected CHO clone 4 cells and transiently transfected CHO clone 29 cells with the confocal microscope of Dr. Baumann of the University of Potsdam (see Results)
- passaging of CHO cells w/ zeocin
- passaging of HT1080
- evaluation of G418 selection experiment: sensitivity of CHO cells towards G418
- transfection of HEK293 and HeLa cells with scFv construct (clone 4) and Nanobody construct (clone 29)
- change of medium in 75cm² flasks with stably transfected CHO Clone 29 (+550µg/ml Hygromycin)
analyzing sequencing results of ligated psB1C3-gene1, 4, 5 constructs
Investigators: Maria, Sascha
Materials:
- sequencing results of GATC from psB1C3-gene1, 4, 5 constructs
- Geneious
Results:
- all sequencing results show complementarity with theoretical gene constructs created in geneious
2012-09-21
cell culture
Investigator:Kerstin
Topic: cell-culture
- cell sorting at the FACS facility of the Deutsches Rheuma-Forschungszentrum (DRFZ): cells were sorted for cells successfully co-transfected with Clone 4 (YFP) and modified AID (GFP) and for cells co-transfected with Clone 29 (mCherry) and modified AID (GFP)
- Immunfluorescence: stably transfected CHO Clone 4 cells were fixed in Ibidi Dishes with 4% PFA for 20min and permeabilized with 0,5% Triton X-100 in PBS for 10 min. The wells were blocked with 1% BSA in PBS for 30min. Incubation with primary antibody (Anti-Flag® M1 Monoclonal Antibody from SIGMA) followed over night at 4°C.
- passaging of HEK293 cells and seeding of 10cm dishes for production of virus
- capturing images of transiently transfected HeLa and HEK293 cells (transfected with Clone 4 and 29)(see Results)
- planning of experiments that should be carried out before the 26. September
- change of medium in 96-well plates with CHO clone 4 cells (+ 550µg/ml Hygromycin)
- collecting supernatant of CHO cells co-transfected with Clone 29 and Cre Recombinase
analyzing sequencing results of ligated psB1C3-gene 2, 3 and -nanobody constructs
Investigators: Maria, Sascha
Materials:
- sequencing results of GATC from psB1C3-gene2, 3 and - nanobodyconstructs
- Geneious
Results:
- all sequencing results show complementarity with theoretical gene constructs created in geneious
2012-09-22
Biobrick glycerol stock, inoculation of o.n.-culture
Investigators: Maria
Materials:
- 12x 15 ml inoculation tubes
- LB-Medium
- Chloramphenicol
- bunsen burner
Method:
- 12x glycerol stock of Biobrick gene1,4,5,6 scFv only, scFv construct
- each o.n.-culture 5ml LB-Medium + 5µl Chloramphenicol
- incubation o.n. at 37°C
further Tasks:
- miniprep of o.n.-cultures
Transformation of Biobrick gene 2 and 3 into new XL1-blue competent E. coli cells
Investigators: Maria
Materials:
- Bunsen Burner, Agar Plate with Chloramphenicol, 37 °C thermo mixer, centrifuge,
- Biobrick gene 2 and 3
- icebox
- new competent E. coli cells (XL 1)
Method:
- according to manual
- 20µl of resuspended cell-suspension were plated on a LB-CM-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones for inoculation of o.n culture
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO cells w/ zeocin and w/o zeocin
- continuation of Immunfluorescence: cells were washed three times with PBS/PBS-Triton and then incubated with secondary antibody (Pacific Blue™ goat anti-mouse IgG (H+L) from life technologies) for two hours. After further washing steps pictures were taken with the fluorescence microscope. (see Results)
- planning of Western Blot for detection of scFv and Nanobody in the cells
2012-09-23
Mini Prep of Biobrick gene 1, 4, 5, 6, scFv only, scFv construct
Investigator:Maria
Materials: overnight cultures Biobrick gene 1, 4, 5, 6, scFv only, scFv construct, Mini prep kit
Methods: according to manual
Results:
scFv only
Gene 1 a: 355,1 ng/µl
Gene 1 b: 351,9 ng/µl<br<
Gene 4 a: 313,9 ng/µl
Gene 4 b: 340,5 ng/µl<br<
Gene 5 a: 346,5 ng/µl
Gene 5 b: 354,8 ng/µl<br<
Gene 6 a: 426,1 ng/µl
Gene 6 b: 478,0 ng/µl<br<
Gene scFv only a: 360,5 ng/µl
Gene scFv only b: 303,4 ng/µl<br<
Gene scFv construct a: 435,3 ng/µl
Gene scFv construct b: 250,6 ng/µl<br<
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of HT1080
- dilution of stably transfected cells (Nanobody construct clone 29, originally from transfected Well 3) to 1-2 cells per 150µl and seeding in 4x 96-Well plates
- seeding of stably transfected CHO Clone 29 cells in Ibidi Dish
- change of medium in 75cm² flasks with stably transfected cells (Clone 4 and 29)
- harvest cells for Western Blot (CHO not transfected, CHO stably transfected Clone 4 and 29)
planning primer for integration of new RAGE-transmembrane domain and RAGE-signalpeptide
Investigator: Sascha
Materials and Methods:
- Geneious
- paper
2012-09-24
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO cells w/ zeocin and w/o zeocin
- seeding CHO cells in Ibidi Dish as control for immunfluorescence experiment with stably transfected Clone 29 cells (planned for 25.09.12)
- passaging HEK 293
Topic: Primer design and ordering
Investigator: Sascha
Materials and Methods: Geneious
Results:
- primerI-fw:rage-signal, xbaI,
- GCCTCTAGAGCTAGCATGGCAGCCGGAACAGCAGTTGGAGCCTGGGTGCTGGTCCTCAGTCTGTGGGGGGCAGTAGTAGGTGCTGACTAC
AAAGACGAAGTGCAATTGCAG
- primerII-rv:c-terminus scfv-te
- GGGCCAGGGCGAGGGTGCCGCTTCCGCCGCCACCGCTTCCCCCTTGAAAATATAAATTCTCTCCAGATCCCCGTTTGATCT
- primerIII-fw:n-terminus of RAGE
- ACCCTCGCCCTGGCCCTGGGAATCTTGGGCGGCCTGGGGACCGCTGCCCTCTTG
- primerIV-rv:c-terminus of RAGE
- CCTTCTCTGCCACAGGATCACGCCGATCAAGAGGGCAGCGGTCCC
- primerV-fw:nterminus of eyfp w
- CGTGATCCTGTGGCAGAGAAGGTCTGGAGTGAGCAAGGGCG
- primerVI: c-terminus of eyfp w
- GCGGCCGCTACTAGTAGGGCCCTTACTTGTACAGCTCGTC
2012-09-25
SDS-PAGE of sonified CHO cells transfected with pcDNA5FRT geneart-nanobody- and scFv-TMD-EYFP constructs
Investigator:Sascha
Materials:
- TEV-protease
- Rotiload SDS-laoding buffer
- heatblock
- SDS-PAGE equipment, TEMED, APS
Method:
- SDS-PAGE Laemli-protocol
2012-09-26
Western Blot (of SDS-PAGEs) of sonified CHO cells transfected with pcDNA5FRT geneart-nanobody- and scFv-TMD-EYFP constructs
Investigator:Sascha
Materials:
- Ponceau-solution
- Whatman paper* PVDF membranes
- Blotting buffer
- Rotiblock
- TBS, TBS+T
- semi-dry plot equipment
- primary antibodies: anti-Flag-tag mouse antibody to detect scFv/scFv construct; anti-human Fc-antibody labeled with biotin
- secondary antibodies: antimouse antibody labeleld with HRP to detect anti-Flag-antibody;streptavidin-HRP conjugat to detect biotinylated anti-Fc-antibody
- ECL(SuperSignal West Pico Trial Kit); Thermo Scientific)
Method:
- Western Blot, semi-dry
Results:
- scFv – after Ponceau staining:
- nanobody – after Ponceau staining:
Topic: Primer design and ordering
Investigator: Sascha
Materials and Methods: Geneious
Results:
- new_primerVI: c-terminus of ey
- GAGCTGCAGCGGCCGCTACTAGTAGGGCCCTTACTTGTACAGCTCGTC
- GAGCTGCAGCGGCCGCTACTAGTAGGGCCCTTACTTGTACAGCTCGTC
2012-09-28
PCR: integration of new RAGE-transmembrane domain and RAGE-signalpeptide into scFv-Tmd-EYFP-construct
Investigators: Sascha
Materials:
- Template: scFv-only Biobrick
- Phusion-polymerase
- 10x Phusion buffer HF
- dNTPs (10mM)
- primerI-fw:rage-signal, xbaI, primerII-rv:c-terminus scfv-te
- Thermocycler
Methods: 3x master mix
| reagent | volume [µL] |
| 10x Phusion HF buffer | 30 |
| dNTPs | 3 |
| primerI-fw:rage-signal td> | 7,5 |
| primerII-rv:c-terminus scfv-te | 7,5 |
| template scFv-only Biobrick (5ng/µl) | 3 |
| Phusion Polymerase | 1,5 |
| water | 97,5 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 15 |
| annealing | gradient: 60;65;70 | 12 | 15 |
| elongation | 72 | 14 | 15 |
| denaturation | 98 | 8 | 15 |
| elongation | 72 | 20 | 15 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
PCR: amplification of EYFP with overhang of new RAGE-TMD
Investigators: Sascha
Materials:
- Template: EYFP_Bba_E0030
- Phusion-polymerase
- 10x Phusion buffer HF
- dNTPs (10mM)
- primerV-fw:nterminus of eyfp w; primerVI: c-terminus of eyfp w
- Thermocycler
Methods: 3x master mix
| reagent | volume [µL] |
| 10x Phusion HF buffer | 30 |
| dNTPs | 3 |
| primerV-fw:nterminus of eyfp w td> | 7,5 |
| primerVI: c-terminus of eyfp w | 7,5 |
| template EYFP (20ng/µl) | 3 |
| Phusion Polymerase | 1,5 |
| water | 97,5 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 30 |
| annealing | gradient: 60;65;69 | 13 | 30 |
| elongation | 72 | 14 | 30 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: analytical gelelectrophoresis of amplified eYFP with RAGE-TMD and RAGE-signalpeptide overhangs
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 90ml
- 105V
Results:
- only amplified eYFP showed sufficient bp size
Further Tasks:
- modifying PCR
- new primer design for advanced PCR
Virus
2012-09-02
Topic: preparing viral stocks
Investigator: Kathi
Aim: preparation of the primary virus stock
Materials:
- cell-culture --> HEK AAV 293 transfected cells on 29.08.2012 (with following plasmids: p01, p06, phelper, psb1c3)
- 80 °C freezer
- 37 °C water bath
Method:
- transfer transfected cells (incl. DMEM) to a falcon tube
- 4 times: 10 min freeze the falcon tube (-80 °C) and 15 min unfreezing (37 °C water bath)
- centrifugation (10 min, 10000 g, RT)
- supernatant = virus stock
- sore at -80 °C
Results:
- virus stock → with YFP on the surface and GOI = CFP
Further tasks
- infection
2012-09-03
Topic: picking clones of p10_Psb1c3_CMV_kozak_sortase_myc and p10_Psb1c3_CMV_kozak_sortase
Investigator: Xenia
Materials:
- LB medium
- ampicillin-stock solution (100 mg/mL)
- plates with E. coli XL1 blue with plasmids: p10_psb1c3_cmv_kozak_sortase_myc and p10_psb1c3_cmv_kozak_sortase
Method:
- picking clones with p10_psb1c3_cmv_kozak_sortase_myc and p10_psb1c3_cmv_kozak_sortase
- over night cultur (50 mL LB medium; 37 °C; 300 rpm; 17 hours)
Further tasks:
- endotoxin free midiprep
Topic: transfection
Investigators: Kerstin and Stefan
Aim:
- The plasmids were transfected in HEK-cells with the following plasmids:
- pSB1C: GOI = CFP (1875 ng)
- phelper (1875 ng)
- P01: YFP linked on VP1 (1875 ng)
- p06: VP1 knock out (1875 ng)
- p06: VP1 knock out (1875 ng)
Materials:
- Medium: opti Mem
- transfection reagent: PEI
- 15 µg DNA per each transfection
Methods:
- transfection preparation
- 1875 ng per plasmid; add 200 µL opti MEM per each transfection
- 4.8 * DNA --> 72 µL PEI add 200 µL per each transfection
- three times 10 s mixing
- 2 min incubation at RT
- centrifagation
- add the PEI approach to the DNA approach
- mixing
- 15 min incubation at RT
- add the transfection approach to the HEK AAV 293 cells
Further tasks
- change medium (DMEM) after 16 hours
- after three days virus maturation, preparation of the primary virus stocks
2012-09-04
Topic: exchange medium
Investigator: Kathi
Materials:
- DMEM
- PBS
Methods:
- asprirate the old medium
- wash with PBS
- add new DMEM on the cells
Further tasks:
- virus preparation on 06.09.2012 (three days virus maturation)
Topic: preparation of the plasmids
Investigators: Kathi
Aim: endotoxin free preparation of p10_psb1c3_cmv_kozak_sortase_myc and p10_psb1c3_cmv_kozak_sortase
Materials and Methods:
- endotoxinfree preparation using the Plasmid plus medi kit (QIAGEN)
- the endotoxic free preparations was tested by disgestion and agarose gel electrophoresis
- digestion: 5 µL DNA + 2 µL FD green buffer + 1 µL of each enzym (XbaI and PstI) and 23 µL water (37 °C and 60 min)
- gel electrophoresis: 1% agarose gel, 100 V, 60 min
Results:
- concentration:
- p10_PsB1c_CMV_kozak_sortase_myc: c= 314,6 ng/µL v=~100 µL
- p10_PsB1c_CMV_kozak_sortase: c= 334,2 ng/µL v=~100 µL
- gel electrophoresis
- the plasmid is wrong – wrong sizes of the plasmids fragments
further tasks
- new preparation of the plasmids
Topic: Infection
Investigators: Kathi
Aim: infection with the virus with YFP on the surface and CFP = GOI; Infected CHO-cells
Materials:
- Virus-stock (prepared on 02.09.2012)
- CHO-cells
- opti HEM
Methods:
- the virus stock was diluted with opti MEM (1:2; 1:10)
- the CHO cells was infected with the same volume of virus solution with the following dilution: 1:1; 1;2; 1;10
- negative control: without virus stock
Further tasks
- microscopy of the infected CHO-cells after 24 and 48 hours
Topic: picking clones of p10_Psb1c3_CMV_kozak_sortase_myc and p10_Psb1c3_CMV_kozak_sortase
Investigator: Xenia
Materials:
- LB medium
- ampicillin 100 mg/mL stock solution
plates with E. coli XL1 blue with plasmids: p10_psb1c3_cmv_kozak_sortase_myc and p10_psb1c3_cmv_kozak_sortase<b
Method:
- picking clones with p10_psb1c3_cmv_kozak_sortase_myc and p10_psb1c3_cmv_kozak_sortase
- over night culture (50 mL LB medium; 37 °C; 300 rpm; 17 hours)
Further tasks:
- endotoxin free midiprep
2012-09-05
Topic: Planing vector plasmid with neomycin cassete
Investigators: Xenia
Aim: planing how to digest and ligate the vectors for the vector plasmid with neomycin cassette
Material and Methods: Geneious
Results:
- pSB1C3 with lITR: cut with SpeI and PstI
- pcDNA3(+) with neomycin cassette: cut with XbaI and PstI
- pSB1C3 with lITR_CMV_neomycin_hGH: cut with SpeI and PstI
- pSB1C3 with rITR: cut with XbaI and PstI
Further tasks:
- design and ordering primer
- practical part:
Topic: preparation of the plasmids
Investigator: Kathi
Aim: endotoxin free preparation of p10_psb1c3_cmv_kozak_sortase_myc and p10_psb1c3_cmv_kozak_sortase
materials and methods:
- endotoxinfree preparation using the medi plasmid kit (QIAGEN)
- the endotoxinfree preparations was tested by digestion and agarose gel electrophoresis
- digestion: 10 µL DNA (1:10 diluted) + 2 µL Green buffer + 1 µL of each enzym (XbaI and PstI) and 17 µL water (37 °C and 60 min)
- gel electrophoresis: 1% agarose gel, 100 V, 60 min
Results:
- concentration:
- p10_PsB1c_CMV_kozak_sortase_myc: c= 1462 ng/µL v=~200 µL
- p10_PsB1c_CMV_kozak_sortase: c= 1350 ng/µL v=~200 µL
- gel electrophoresis
- the plasmid is wrong – wrong sizes of the plasmids fragments
- the wrong antibiotic was used in the over night culture
Further tasks
- retransformation of the right p10_psb1c_CMV kozak_sortase_myc and
p10_psb1c_CMV kozak_sortase plasmids
Topic: transfection
Investigator: Kathi
Aim:
- The plasmids were transfected in HEK-cells with the following plasmids:
- pSB1C: GOI is CFP (1875 ng)
- phelper (1875 ng)
- P10_PsB1c3_CMV_kozak_sortase_myc (1875 ng)
- p06: VP2 knock out (1875 ng)
- p06: VP2 knock out (1875 ng)
- The plasmids were transfected in HEK-cells with the following plasmids:
- pSB1C: GOI is CFP (1875 ng)
- phelper (1875 ng)
- P10_PsB1c3_CMV_kozak_sortas (1875 ng)
- p06: VP2 knockut 1875 ng)
- p06: VP2 knockut 1875 ng)
Materials:
- PEI (transfection reagent)
- opti MEM (transfection medium)
- endotoxin free preparated plasmids
Further tasks
- change medium (DMEM) after 16 hours
- after three days virus maturation – preparation of the virus
2012-09-06
Topic: Primer design and ordering
Investigator: Xenia
Materials and Methods: Geneious
Results:
- Primer (forward) with XbaI recognition site:
- GTCTAGACACGGAATGTGTGTCAGTTAGGGTGTGG
- Primer (reverse, complement) with SpeI and PstI recognition site:
- TACTGCAGCCCCTACTAGTATCGGGCAGACATGATAAGATACATTGATGAGTTTGG
Annealing temp.: 71 °C
Topic: exchange medium
Investigator: Kathi
Materials:
- DMEM
- PBS
Methods:
- asprirate the old medium
- wash with PBS
- add new DMEM to the cells
Further tasks:
- virus preparation on 09.09.2012 (four days virus maturation)
Topic: preparing viral stocks
Investigator: Kathi
Aim: virus preparation; virus constuct: YFP on the surface and GOI=CFP
Materials:
- cell-culture --> HEK AAV 293 transfected cells on 03.09.2012 (with following plasmids: p01, p06, phelper, psb1c3)
- 80 °C freezer
- 37 °C water bath
Method:
- transfer transfected cells (incl. DMEM) to a falcon tube
- 4 times: 10 min freeze the falcon tube (-80 °C) and 10 min unfreezing (37 °C water bath)
- centrifugation (10 min, 10000 g, RT)
- supernatant = virus stock
- sore at -80 °C
Results:
- virus stock → with YFP on the surface and GOI = CFP
Further tasks
- infection of the CHO-cells
Topic: microscopy
Investigators: Stefan and Kerstin
<b> Aim: virus preparation
Materials and methods:
- microscopy of the infeced CHO cells (02.09.2012)
Results:
- there are yellow fluorescence particle visible
- after bleaching there arn´t yellow fluorescence particle vibible
Further tasks
- another infection with a positive control (infected HT1080 cells)
2012-09-09
Topic: preparing viral stocks
Investigator: Kathi
Aim: virus preparation
Materials:
- cell-culture --> HEK AAV 293 transfected cells on 05.09.2012 (with following plasmids: p10_PsB1c_CMV_kozak_sortase_myc, VP2 knock out, phelper, PsB1c3GOI=CFP and p10_PsB1c_CMV_kozak_sortase, VP2 knock out, phelper, PsB1c3GOI=CFP)
- 80 °C freezer
- 37 °C water bath
Method:
- transfer transfected cells (incl. DMEM) to a falcon tube
- 4 times: 10 min freeze the falcon tube (-80 °C) and 15 min unfreezing (37 °C water bath)
- centrifugation (10 min, 10000 g, RT)
- supernatant = virus stock
- sore at -80 °C
Results:
- virus stock → surface: sortase + myc-tag; GOI= CFP
virus stock → surface: sortase; GOI= CFP
- virus stock → surface: sortase; GOI= CFP
virus stock → surface: sortase; GOI= CFP
Further tasks
- infection
2012-09-10
Topic: re-transformation
Investigator: Kathi
Materials:
- LB medium
- competent cells (XL1-blue cells)
Method:
- transformation via manual, 1 µL of the sample (PsB1c3_CMV_kozak_sortase_myc and PsB1c3_CMV_kozak_sortase prepared on 17.08.2012) were used
Further tasks:
- picking clones
2012-09-11
Topic: preparation of the plasmids
Investigator: Kathi
Aim:endotoxinfree preparation of the following plasmids: GOI=CFP, CFP linked on VP2 and GOI=mVenus
Materials and Methods:
- endotoxinfree preparation using the plasmid plus medi kit (QIAGEN)
Results:
- concentration:
- GOI=CFP – c= 1159 ng/µL; v= 200 µL
- CFP linked on VP2 – c= 776 ng/µL; v= 200 µL
- GOI=mVenus – c= 532 ng/µL; v= 200 µL
Further tasks
- transfection with GOI= mVenus and CFP linked on VP2
Topic: PCR
Investigator: Xenia
Aim: PCR-amplificate: Neomycin cassette with restriction sites XbaI as prefix and SpeI and PstI as suffix
Materials:
- PCR
- Vektor with antibiotic resistance: pcDNA3(+)
- Primer:
- r_primer2_ neomycin_cassette-spe_pst (reversed):GTCTAGACACGGAATGTGTGTCAGTTAGGGTGTGG
- f_primer_neomycin_cassette_xbaI_fertig:TACTGCAGCCCCTACTAGTATCGGGCAGACATGATAAGATACATTGATGAGTTTGG
- Phusion Polymerase, dNTPs, HF buffer
- Gel elektrophorese
- Nucleo Spin Gel and PCR Cleanup (Macherey-nagel)
Method:
- polymerase chain reaction
Mastermix
| reagent | volume [µL] |
| HF Phusion buffer 5x | 10 |
| dNTPs | 1 |
| Primer (Forward) | 2 |
| Primer (Reverse) | 2 |
| DNA (Plasmid) | 1.0 |
| Phusion Polymerase | 0.5 |
| water | 33.5 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 10 | 30 |
| annealing | 71 | 40 | 30 |
| elongation | 72 | 40 | 30 |
| final elongation | 72 | 300 | 1 |
| cooling | 8 | ∞ | 1 |
Results:
The concentration of the template was too high (1 µg/µL). The digestion couldn't be done because the template contains the same restriction sites as the PCR-amplificate
Further tasks:
- PCR with lower template concentration
- Gel electophorese
- Digestion
Topic: PCR
Investigator: Xenia
Aim: PCR-amplificate: Neomycin cassette with restriction sites XbaI as prefix and SpeI and PstI as suffix
Materials:
- Vektor with antibiotic resistance: pcDNA3(+)
- Primer:
- r_primer2_ neomycin_cassette-spe_pst (reversed):GTCTAGACACGGAATGTGTGTCAGTTAGGGTGTGG
- f_primer_neomycin_cassette_xbaI_fertig:TACTGCAGCCCCTACTAGTATCGGGCAGACATGATAAGATACATTGATGAGTTTGG
- Phusion Polymerase, dNTPs, HF buffer
- Gel elektrophorese
- NucleoSpin Gel ans PCR Clean-up Kit (macherey-nagel)
Method:
- polymerase chain reaction
Mastermix
| reagent | volume [µL] |
| HF Phusion buffer 5x | 10 |
| dNTPs | 1 |
| Primer (Forward) | 2 |
| Primer (Reverse) | 2 |
| DNA (Plasmid) | 1.0 |
| Phusion Polymerase | 0.5 |
| water | 33.5 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 10 | 30 |
| annealing | 71 | 40 | 30 |
| elongation | 72 | 40 | 30 |
| final elongation | 72 | 300 | 1 |
| cooling | 8 | ∞ | 1 |
Further tasks:
- Gel electophorese
- Digestion
2012-09-12
Topic: transfection
Investigator: Kerstin
Aim:
- The plasmids were transfected in HEK-cells with the following plasmids:
- CFP linked on VP2 (1875 ng)
- VP2 knock out (1875 ng)
- phelper (1875 ng)
- PsB1c3-GOI=CFP (1875 ng)
- PsB1c3-GOI=CFP (1875 ng)
Materials:
- PEI (transfection reagent)
- opti MEM (transfection medium)
- endotoxin free preparated plasmids
Further tasks
- change medium (DMEM) after 16 hours
- after three days virus maturation – preparation of the virus (14.09.2012)
Topic: Gelelectrophoresis with PCR-amplificate
Investigator: Xenia
Aim: check whether the PCR worked
Materials:
- Geneious
- agarose
- 1x TAE-buffer
- 10x FD green Buffer
- PCR-sample
Method:
Gelelectrophoresis of PCR Neomycin cassette (1533bp) uncut
Results:
No band indicating template is visible. The neomycin cassette will be cut.
further tasks:
- purification with PCR-clean up
- Digestion of neomycin cassette
Topic: Purification of PCR-amplificate with PCR clean-up Kit
Investigator: Xenia
Aim:
Purification of PCR-amplificate
Materials:
- NucleoSpin Gel and PCR Clean-up Kit (macherey-nagel)
Results:
- concentration of amplified neomycin cassette: 106.2 ng/µL
Further tasks:
- Digestion of amplified neomycin cassette
Topic: purification of p10-pSB1C3-CMV-Kozak-Sortase
Investigator: Xenia
Aim: purification of p10-pSB1C3-CMV-Kozak-Sortase
Materials:
Plasmid Plus Midi Kit (QIAGEN)
Results:
concentration of plasmid:
- p10-pSB1C3-CMV-Kozak-Sortase: 2045.8 ng/µL
Further tasks:
- transfection
Topic: purification of left and right ITR plasmids with miniprep
Investigator: Xenia
Aim: purification of left and right ITR plasmids
Materials:
- Geneious
- GeneJET Plasmid Miniprep Kit
- Gel electrophoresis
Results:
concentration of the plasmids:
- left ITR: 174.4 ng/µL
- left ITR: 224.2 ng/µL
- right ITR: 87.2 ng/µL
- right ITR: 75.5 ng/µL
Gel electrophoresis
The left_ITR plasmid consists of 2.217 kbp and right ITR consists of 2.216 kbp
Further tasks:
digestion
send the DNA to sequencing
Investigators: Xenia and Tobias
| sample | GATC number | Seq. Primer |
| p10-pSB1C3-CMV-Kozak-Sortase | II3673 | p140:Viral Brick 587KO-His rev |
| p10-pSB1C3-CMV-Kozak-Sortase | II3674 | p37: Primer cap 2800 for |
| p10-pSB1C3-CMV-Kozak-Sortase | II3675 | p39: Primer cap 3500 for |
| p10-pSB1C3-CMV-Kozak-Sortase | standard Primer GATC | CMV-Primer |
Topic: EGFR - resuspend EGFR construct
Investigator: Tobi
Aim: resuspend of the EGFR construct
Materials and Methods:
- resuspend the EGFR in 50 µL water
Further tasks:
- transformation
Topic: EGFR - transformation
Investigator: Tobi
Aim: transformation of the EGFR construct
Materials and Methods:
- LB-medium
- competent cells (XL1-blue cells)
- transformation via manual, 10 µL of the ligation were used
Further tasks:
- picking clones
2012-09-13
Topic: Virus preparation - transfection
Investigator: Kathi
Aim: transfection of HEK-AAv 293 cells -> virus contruct with kozak_sortase_myc + GOI=CFP and kozak_sortase + GOI=CFP
Materials:
- plasmids:
- p10_psb1c_CMV_kozak_sortase_myc (1875 ng)
- p05 - VP2 knock out (1875 ng)
- GOI = CFP (1875 ng)
- phelper (1875 ng)
- PEI (transfection reagent)
- opti MEM (transfection medium)
Methods:
- transfection preparation1875 ng per plasmid
- add 200 µL opti MEM-medium per each transfection
- 4.8 * DNA --> 72 µL PEI add 200 µL per each transfection
- three times 10 s mixing
- 2 min incubation at RT
- centrifagation
- add the PEI preparation to the DNA preparation
- mixing
- 15 min incabation at RT
- add the transfection preparation to the HEK AAV 293 cells
Further tasks:
- change medium (DMEM) after 16 hours
- after three days virus maturation – preparation of the virus (16.09.2012)
Topic: Virus preparation - exchange medium
Investigator: kathi
Aim: change the medium of the transfected HEK cells (transfection date: 12.09.2012)
Materials:
- PBS
- DNEN
Methods:
- asprirate the old medium
- wash with PBS
- add new DMEM to the cells
Further tasks:
- virus preparation on 17.09.2012 (three days virus maturation)
Topic: p10_psb1c_CMV_kozak_sortase - +myc and - myc - Digestion
Investigator: Kathi
Aim: test digestion
Materials:
- plasmids:
- p10_psb1c_CMC_kozak_sortase_myc
- p10_psb1c_CMC_kozak_sortase
- enzymes: XbaI and PstI
- 10 x FD green buffer
Methods:
- Digestion approache:
- 5 µL DNA (1:100 dilute) + 1 µL of each enzyme + 3 µL 10 x FD green buffer + 21 µL water
- 1 hour at 37 °C
Further tasks:
- gel electrophoresis
Topic: p10_psb1c_CMV_kozak_sortase - +myc and - myc - gel-electrophoresis
Investigator: Kathi
Aim: control of the test digest
Materials:
- 1% agarose gel
- 1 x TAE buffer
Methods:
- run: 60 min at 100 V
Further tasks:
- sequencing
Topic: EGFR - picking clones
Investigator: Tobias
Aim: picking two clones
Materials and Mtehods:
- picking two clones and incubate over night in LB medium (incl. Amp)
Further tasks:
- DNA preparation
2012-09-14
Topic: Sequencing
Investigator: Kathi
Aim: send the re-tranfected p10_psb1c3_CMV_kozak_sortase_myc and p10_psb1c3_CMV_kozak_sortase for sequncing
Materials:
- Primer
- concentration and volum of the DNA: c= 50 ng/µL; v= 100 µL
Methods: GATC
Further tasks:Geneious analysis
Topic: exchange medium
Investigator: Kathi
Aim: change the medium of the transfected HEK cells (transfection date: 13.09.2012)
Materials:
- PBS
- DMEM
Methods:
- asprirate the old medium
- wash with PBS
- give new DMEM on the cells
Further tasks:
Topic: infection
Investigator: Kerstin
Aim: infection of the HT1080, CHO-cells and CHO-cells with nanobody
Materials and Methods:
- the virus was added to the cells
Further tasks:
- aspriration of the medium (inkl. virus) and add of new virus-medium mix
Topic: transfection
Investigator: Kathi
Aim: transfection of HEK-AAv 293 cells -> virus contruct with CFP on the surface and GOI= mVenus
Materials
- plasmids:
- CFP linked on VP2 (1875 ng)
- VP2 knock out (1875 ng)
- GOI = mVenus (1875 ng)
- phelper (1875 ng)
- PEI (transfection reagent)
- opti MEM (transfection medium)
Methods:
- transfection preparation1875 ng per plasmid; add 200 µL opti MEM-medium per each transfection
- 4.8 * DNA --> 72 µL PEI add 200 µL per each transfection
- three times 10 s mixing
- 2 min incubation at RT
- centrifagation
- add the PEI preparation to the DNA preparation
- mixing
- 15 min incabation at RT ä
- add the transfection preparation to the HEK AAV 293 cells
Further tasks:
- change medium (DMEM) after 16 hours
- after three days virus maturation – preparation of the virus (17.09.2012)
Topic: EGFR - DNA preparation
Investigator: Kathi
Aim: preparation of the EGFR over night culture
Materials and Methods:
- over night culture
- GeneJET Plasmid Miniprep Kit
Results:
- concentration
- EGFR - clon 1: c= 321 ng/µL v= 50 µL
- EGFR - clon 2: c= 383 ng/µL v= 50 µL
Further tasks:
- Digestion
Topic:EGFR - preparatively digestion
Investigator: Kathi
Aim: digest the EGFR construct with XbaI and AgeI for the BioBrick and with XbaI and PstI for the arabinose plasmid
Materials:
- 10 x FD green buffer
- DNA (preped with JetGen-Kit)
- XbaI, AgeI and PstI
Methods:
- EGFR-construct clon 1:
- degestion approach (BioBrick): 6,2 µL DNA (~ 2 µg DNA) + 2 µL of each enzyme (XbaI and AgeI) + 3 µL FD green buffer + 18,8 µL water
- degestion approach (Arabinose): 6,2 µL DNA (~ 2 µg DNA) + 2 µL of each enzyme (XbaI and PstI) + 3 µL FD green buffer + 18,8 µL water
- EGFR-construct clon 2:
- degestion approach (BioBrick): 5,2 µL DNA (~ 2 µg DNA) + 2 µL of each enzyme (XbaI and AgeI) + 3 µL FD green buffer + 19,8 µL water
- degestion approach (Arabinose): 5,2 µL DNA (~ 2 µg DNA) + 2 µL of each enzyme (XbaI and PstI) + 3 µL FD green buffer + 19,8 µL water
- 2,5 hours; 37 °C
Further tasks:
gel electrophoresis
Topic: EGFR - gel-electrophoresis
Investigator: Kathi
Aim: seperation of the fragments
Materials and Methods:
- 1% agarose
- 1 x TBE buffer
- 90 min at 95 V
Results:
Further tasks:
gel extraction
Topic: EGFR - ligation
Investigator: Kathi
Aim: ligate the EGFR construct into the BioBrick plasmid (Backbone) and the arabinose plasmid
Materials:
- T4 ligase
- T4 ligase buffer
Methods:
- ligase approach:
- EGFR-BioBrick:
- 2 µL Ta ligase buffer + 1 µL ligase + 1.2 µL psb1cplasmid (backbone) + 6.9 µL EGFR construckt (digest with XbaI, AgeI)
- EGFR-agabinose:
- 2 µL Ta ligase buffer + 1 µL ligase + 3.7 µL arabinose plasmid (backbone) + 3.3 µL EGFR construckt (digest with XbaI, PstI)
Further tasks:
transformation
Topic: EGFR - transformation
Investigator: Kathi
Aim: transformation - EGFR construct/BioBrick and EGFR/arabinose
Materials:
- LB-medium
- competent cells (XL1-blue cells)
- antibiotics
- arabinose backbone - Amp
- BioBrick backbone - CM
Methods:
- transformation via manual, 10 µL of the ligation were used
Further tasks:
picking clones
Topic: lITR-antibiotic-rITR - picking clones
Investigator: Kathi
Aim: picking clones - over nicht clutute
Materials and Methods:
- LB-medium with CM
Further tasks:
DNA peparation
2012-09-15
Topic: Virus-construct - Virus preparation
Investigator: kathi
Aim: preparing the primary virus stock --> CFP on the surface anf GOI = mVenus
Materials:
- cell-culture --> HEK AAV 293 transfected cells on 14.09.2012
- 80 °C freezer
- 37 °C water bath
Methods:
- transfer transfected cells (incl. DMEM) into a falcon tube
- 4 times: 10 min freeze the falcon tube (-80 °C) and 15 min unfreezing (37 °C water bath)
- centrifugation (10 min, 10000 g, RT)
- supernatant = virus stocksore at -80 °C
Results:
- primary virus stock with CFP on the surface and mVenus = GOI
Further tasks:
- infection
Topic: exchange medium
Investigator: Kathi
Aim: Change the medium of the transfected HEK cells (transfection date: 14.09.2012)
Materials:
- PBS
- DMEM
Methods:
- asprirate the old medium
- wash with PBS
- add new DMEM to the cells
Further tasks:
- virus preparation on 17.09.2012 (three days virus maturation)
Topic: EGFR - picking clones
Investigator: kathi
Aim: picking clones - the EGFR contruct into the arabinose and psb1c plasmid
Materials and Methods:
- LB-medium
- antibiotics
- arabinose plasmid - Amp
- psb1c plasmid - CM
- over night incubation - shaking - 37 °C
Further tasks:
- arabinose plasmid - mainculter - start expression
- psb1c plasmid - preparation of the DNA
Topic: ITR: preparation
Investigator: kathi
Aim: preparation of the ITR-neo over night cultur
Materials and Methods:
- GeneJET Plasmid Miniprep Kit
Results:
- concentration
- clone 1: c= 335 ng/µL v= 50 µL
- clone 2: c= 325 ng/µL v= 50 µL
Further tasks:
test digestion
Topic: ITR: Digestion
Investigator: kathi
Aim: Digestion of the prapared plasmids (leftITR+neo)
Materials:
- 10 x FD green buffer
- DNA (preped with JetGen-Kit)
- SpeI and PstI
Methods:
- 2 µg DNA + 3 µL FD green buffer + 2 µL of each enzyme + add 30 µL water
Results:
Further tasks:
new digestion with less DNA - 1000 ng and 500 ng
2012-09-16
Topic: Virus-construct - Virus preparation
Investigator: kathi
Aim: preparing the primary virus stock --> p10_psb1c3_CMV_kozak_sortase_myc and p10_psb1c3_CMV_kozak_sortase
Materials:
- cell-culture --> HEK AAV 293 transfected cells on 13.09.2012
- 80 °C freezer
- 37 °C water bath
Methods:
- transfer transfected cells (incl. DMEM) to a falcon tube
- 4 times: 10 min freeze the falcon tube (-80 °C) and 15 min unfreezing (37 °C water bath)
- centrifugation (10 min, 10000 g, RT)
- supernatant = virus stock store at -80 °C
Results:
- primary virus stock
- p10_psb1c3_CMV_kozak_sortase_myc
- p10_psb1c3_CMV_kozak_sortase
Further tasks:
- infection
Topic: EGFR - preparation (BioBrick)
Investigator: kathi
Aim: preparation of the EGFR construct
Materials and Methods:
- over night culture
- GeneJET Plasmid Miniprep Kit
Results:
- concentration EGFR (construct)- clone 1: c= 285 ng/µL v= 50 µL
- concentration EGFR (construct)- clone 1: c= 385 ng/µL v= 50 µL
Further tasks:
- Digestion
Topic: EGFR - Digestion (BioBrick)
Investigator: kathi
Aim: Test Digestion
Materials
- fast digest (FD) enzymes (XbaI and AgeI)
- FD green buffer
Methods:
- Digestion: 5 µL (1:10 diluted DNA) + 3 µL FD green buffer + 1 µL of each enzyme add 30 µL water
Results:
- the bands seems to be correct
Further tasks:
send for sequencing
2012-09-17
Topic: Virus-construct - Virus preparation
Investigator: kathi
Aim: preparing the primary virus stock --> CFP on the surface and GOI= mVenus
Materials:
- cell-culture --> HEK AAV 293 transfected cells on 14.09.2012
- 80 °C freezer
- 37 °C water bath
Methods:
- transfer transfected cells (incl. DMEM) to a falcon tube
- 4 times: 10 min freeze the falcon tube (-80 °C) and 15 min unfreezing (37 °C water bath)
- centrifugation (10 min, 10000 g, RT)
- supernatant = virus stock store at -80 °C
Results:
- primary virus stock
- CFP on the surface and YFP = GOI
Further tasks:
- infection
Topic: EGFR - expression start
Investigator: kathi
Aim: start the expression with arabinose
Materials:
- LB-medium
- arabinose [10%]
- ampicilin
Methods:
- 200 µL over night culture into fresh LB medium (incl. Amp.)v= 150 µL
- incubation till OD 0,4
- start of the expression + 450 µL 10% arabinose solution
- 4 hours incubation 37 °C
- the pellet can be store at -20 °C
Further tasks:
preparation of the EGFR
Topic: qPCR
Investigator: kathi
Aim: test the genetic virus titer of the virus with YFP on the surface and GOI = CFP
Materials:
- 2 x QuantiFast SYBR-green (QIAGEN)
- Rnase free water
- RNaseI (NEB)
- CMV primer
- CMV_forward_qPCR: 5' - GGGACTTTCCTACTTGGCA - 3'
- CMV_reverse_qPCR: 5' - GGCGGAGTTGTTACGACA - 3'
Methods:
- the stard qPDR protocol were used
Results:
- there are no evaluable quantification curves
Further tasks:
- repetition of the experiment
2012-09-18
Topic: ITR-neo Digestion
Investigator: kathi
Aim: digestion of the ITR-neo contruct
Materials and Methods:
- ITR-neo clone 1
- 3 µL DNA (1000 ng)+ 1,5 µL per enzyme (PstI; SpeI) + 3 µL buffer 4 (NEB) 22 µL water
- 1,5 µL DNA (500 ng) + 1,5 µL per enzyme (PstI; SpeI) + 3 µL buffer 4 (NEB) 23,5 µL water
- ITR-neo clone 2
- 3 µL DNA (1000 ng)+ 1,5 µL per enzyme (PstI; SpeI) + 3 µL buffer 4 (NEB) 22 µL water
- 1,5 µL DNA (500 ng) + 1,5 µL per enzyme (PstI; SpeI) + 3 µL buffer 4 (NEB) 23,5 µL water
- 37 °C; 60 min
Results:
Further tasks:
- shorter digestion time
- desing of new primer
Topic: EGFR - send for sequencing
Investigator: kathi
Aim: send the EGFR in the psb1c plasmid for sequencing
Materials and Methods:
- send the EGFR clone 1 and clone 2 (c= 50 ng/µL)
- Primer forward and reverse psb1c primer
Topic: picking clones
Investigator: Xenia
Materials:
- LB medium
- chloramphenicol sock solution (25 mg/mL)
- plates with E. coli with plasmids: EGFR
Method:
- picking clones with EGFR
- over night cultur (50 mL LB medium; 37 °C; 300 rpm; 17 hours)
Further tasks:
- expression of EGFR
2012-09-19
Topic: EGFR - expression start
Investigator: Xenia
Aim: start the expression with arabinose
Materials:
- LB-medium
- arabinose [10%]
- ampicilin
Methods:
- 25 mL overnight culture into fresh 1 L LB medium (incl. Amp.)v= 1 mL
- incubation till OD 0.4
- start of the expression: 5 mL 10% arabinose solution (c=0.05 %)
- 4 hours incubation of clone I by 30 °C and of clone II by 37 °C.
- the pellet can be stored at -20 °C
Results:
- clone I was incubated till OD 0.39 and
- clone II was incubated till OD 0.42
Further tasks:
Western blot: the band should appear by 24,8 kDa
- Gel: 12.5 %
- Sample for western blot: 80 µL Sample (OD ~ 2.5) + 20 µL Roti-Load1 reducing
- denaturation 10 min; 95 °C
2012-09-20
Topic: EGFR - SDS-PAGE
Investigators: Xenia and Kathi
Aim: separation of the EGFR using the SDS PAGE - control of the expresion
Materials:
- 12.5% SDS-gel
- 1 x TBS buffer
Methods:
- load 20 µL of the praparted samples to the SDS gel
- 100 V 1.5 hours
Further tasks:
Western blot: the band should appear by 24,8 kDa
Topic: EGFR - western blot
Investigator: Kathi
Aim: detection of the EGFR
Materials:
- membrane
- TBS-buffer + 5% milk powder
Methods:
- semi-dry plot of the SDS-PAGE
- blocking of the memrane over night
Topic: ITR - PCR clean up
Investigator: Kathi
Aim: PCR clean up of the 0.4 PCR
Materials and Methods:
- NucleoSpin Gel and PCR Clean-up Kit (macherey-nagel)
Further tasks:
- Digestion
Topic: ITR - Digestion
Investigator: Kathi
Aim: contol of the PCR product and the ligation (leftITR-neo)
Materials:
- fast digest enzyms
- fast digest green buffer
- Enzyms: PstI, XbaI, SpeI
Methods:
- leftITR-neo
- Digestion: 1 µL SpeI + 0,3 µL PstI + 3 µL FD green buffer add 30 µL water (three times: 10 min, 20 min and 40 min digestion time)
- PCR
- Digestion: 2 µL XbaI + 2 µL PstI + 3 µL FD green buffer add 30 µL water
- leftITR
- Digestion: 2 µL SpeI + 2 µL PstI + 3 µL FD green buffer add 30 µL water
one hour at 37 °C
Results:
Further tasks:
- new PCR with neu primer
- new ligation with the neu pcr product
2012-09-21
Topic: EGFR - staining of the membrane
Investigators: Xenia and Kathi
Aim: staining of the membrane
Materials:
- TBST buffer + milk powder
- antibody:
Methods:
- washing three times of the membrane with TBST
- 15 min incubation with TBST buffer
- three times washing of the membrane
- incubation with the antibody
- three times washing of the membrane with TBST
- staining of the membraine
Results:
Further tasks:
- periplasma preparation
2012-09-22
Topic: Virus - transfection
Investigator: Kathi
Aim: The plasmids were transfected in HEK-cells with the following plasmids: pSB1C: GOI = CFP (1875 ng) phelper (1875 ng) P01: YFP linked on VP1 (1875 ng) p06: VP1 knock out (1875 ng)
Materials:
- Medium: opti Mem
- transfection reagent: PEI 15 µg per each transfection
Methods:
- transfection preparation 1875 ng per plasmid; add 200 µL opti MEM per each transfection 4.8 * DNA --> 72 µL PEI add 200 µL per each transfection
- three times 10 s mixing
- 2 min incubation at RT
- centrifagation
2012-09-25
Topic: Virus - qPCR
Investigator: Kathi
Aim: test the genomic virus titer of the viruses with YFP on the surface and CFP=GOI
Materials:
- RNase free watreä
- primer
- CMV_forward_qPCR: 5' - GGGACTTTCCTACTTGGCA - 3'
- CMV_reverse_qPCR: 5' - GGCGGAGTTGTTACGACA - 3'
- DNaseI (NEB)
- 2x QuantiFast SYBR-green (QIAGEN)
Methods:
- the real time standard protocol were used
- there are 3,47 copies of the virus in each PCR
</div>
2012-09-29
Topic: PCR - Kanamycin cassette
Investigator: Xenia
Materials:
- Kanamycin resistance cassette
- r_primer2_ neomycin cassette-spe_pst-fertig2: CCAAACTCATCAATGTATCTTATCATGTCTGCCCGATACTAGTAGGGGCTGCAG
- RFC10prefix_f_primer_neomycin_cassette_xbaI_fertig: GAATTCGCGGCCGCTTCTAGACGGAATGTGTGTCAGTTAGGGTGTGG
Methods:
- final concentration of DNA: 200 ng
- Annealing temperature: 71 °C
Further tasks:
Gelelectrophorese or PCR with final concentration of DNA: 20 ng
Topic: PCR - Kanamycin cassette
Investigator: Xenia
Materials:
- Kanamycin resistance cassette
- r_primer2_ neomycin cassette-spe_pst-fertig2: CCAAACTCATCAATGTATCTTATCATGTCTGCCCGATACTAGTAGGGGCTGCAG
- RFC10prefix_f_primer_neomycin_cassette_xbaI_fertig: GAATTCGCGGCCGCTTCTAGACGGAATGTGTGTCAGTTAGGGTGTGG
Methods:
- final concentration of DNA: 20 ng
- Annealing temperature: 71 °C
Further tasks:
Gelelectrophorese
2012-10-16
Topic: Gelelectrophoresis
the first two samples are with 200 ng and the 3rd sample is with 20 ng final concentration of template
2012-10-16
Topic: PCR clean up
 "
"