Team:Paris Bettencourt/Restriction Enzyme
From 2012.igem.org
The combination of plasmids:
- First plasmid: pLac & RBS & I-SceI [Cm]
- Second plasmid: I-SceI restriction site [Amp]
From photos and bar graph above we can see that our expectation came true.
Before to go to Boston, we improved our protocol to characterize TUDelft BioBrisks and showed the result reproducibility.
New protocol:
Both TUDelft plasmids are trasformed to the E.coli NEB Turbo strain:
- First plasmid: High copy plasmid with encoded generator to express I-SceI meganucllease, [http://partsregistry.org/Part:BBa_K175041 BBa_K175041]:
- Resistance: Chloramphenicol
- Resistance: Chloramphenicol
- Second plasmid: High copy plasmid with encoded I-SceI restriction site, [http://partsregistry.org/Part:BBa_K175027 K175027]:
- Resistance: Ampicillin and Kanamycin
- Start a liquid culture with Amp and Cm to select bacteria wich have both plasmids.
- Incubate at 37°C overnight.
- Pellet and wash cells to remove Amp and Cm.
- Re-dilute them in the LB to have OD = 0.05.
- From the tube, start two liquid cultures:
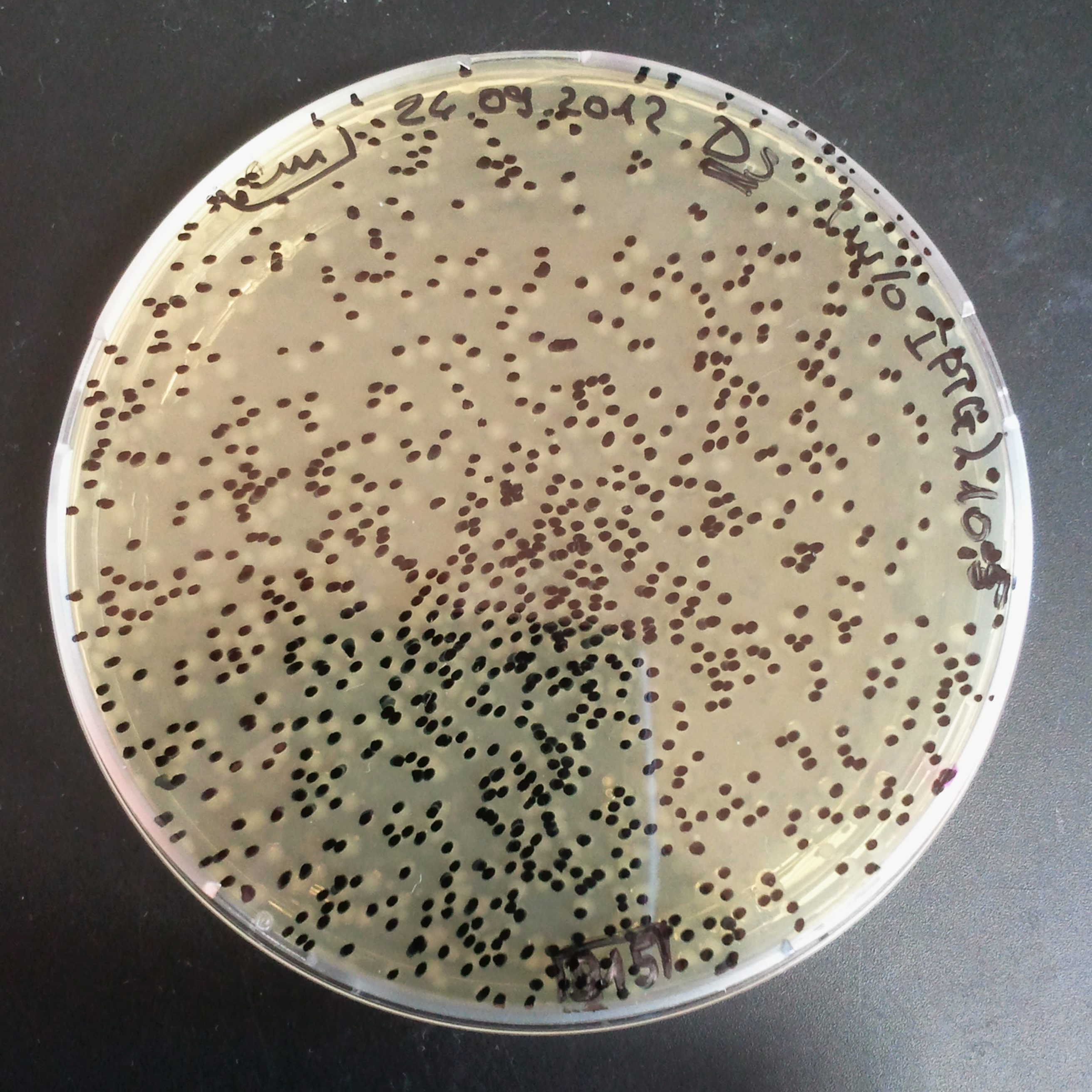
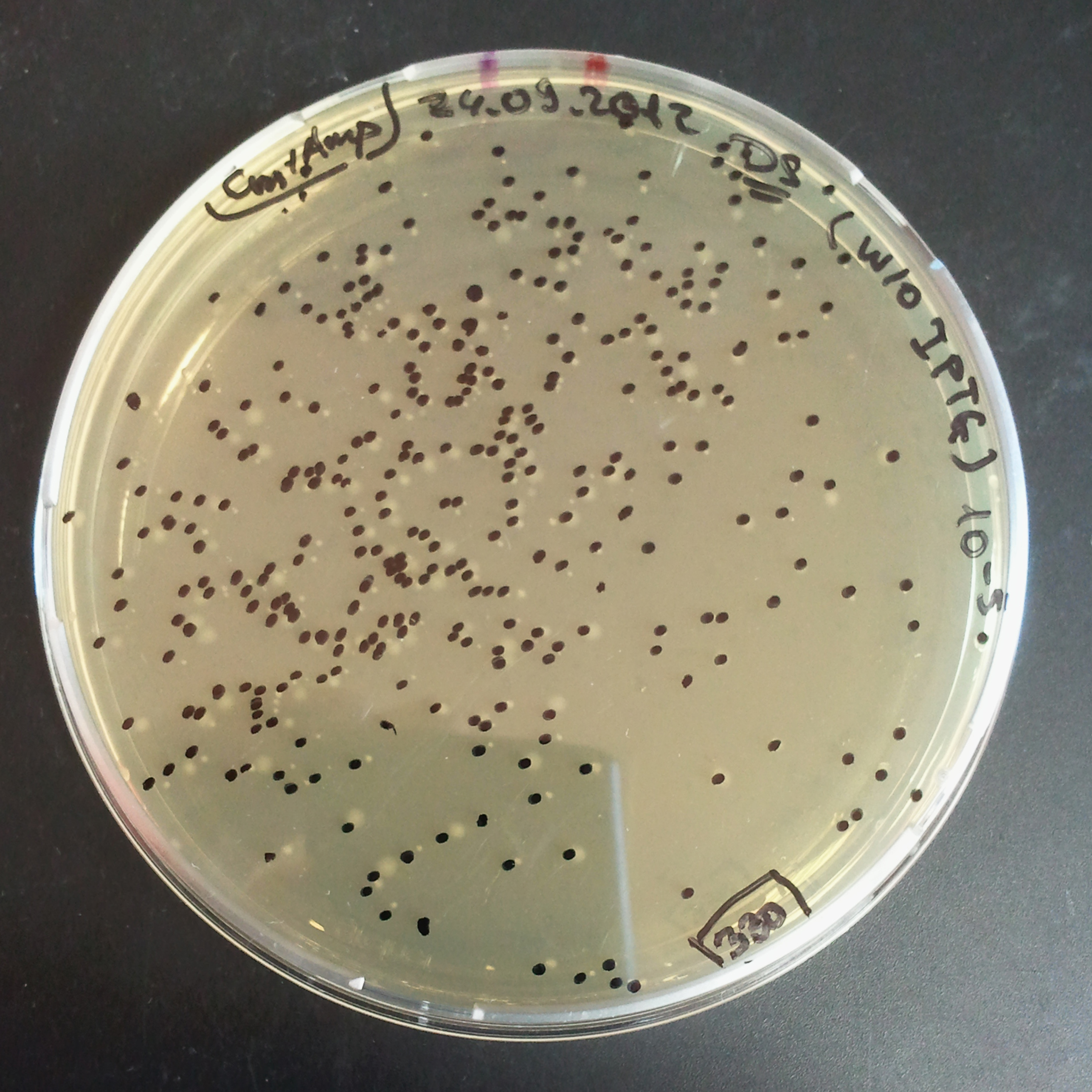
- The first culture with Cm and w/o IPTG.
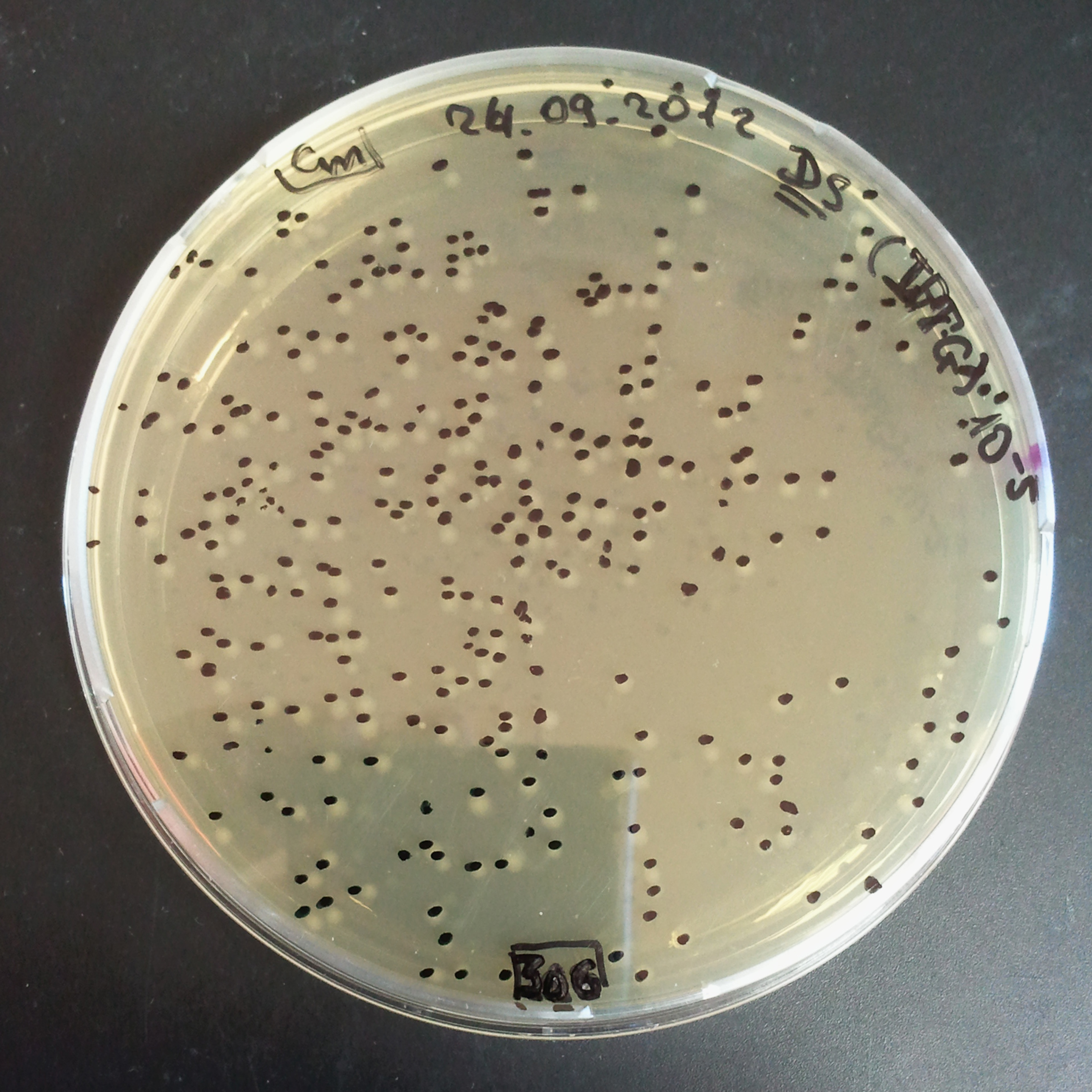
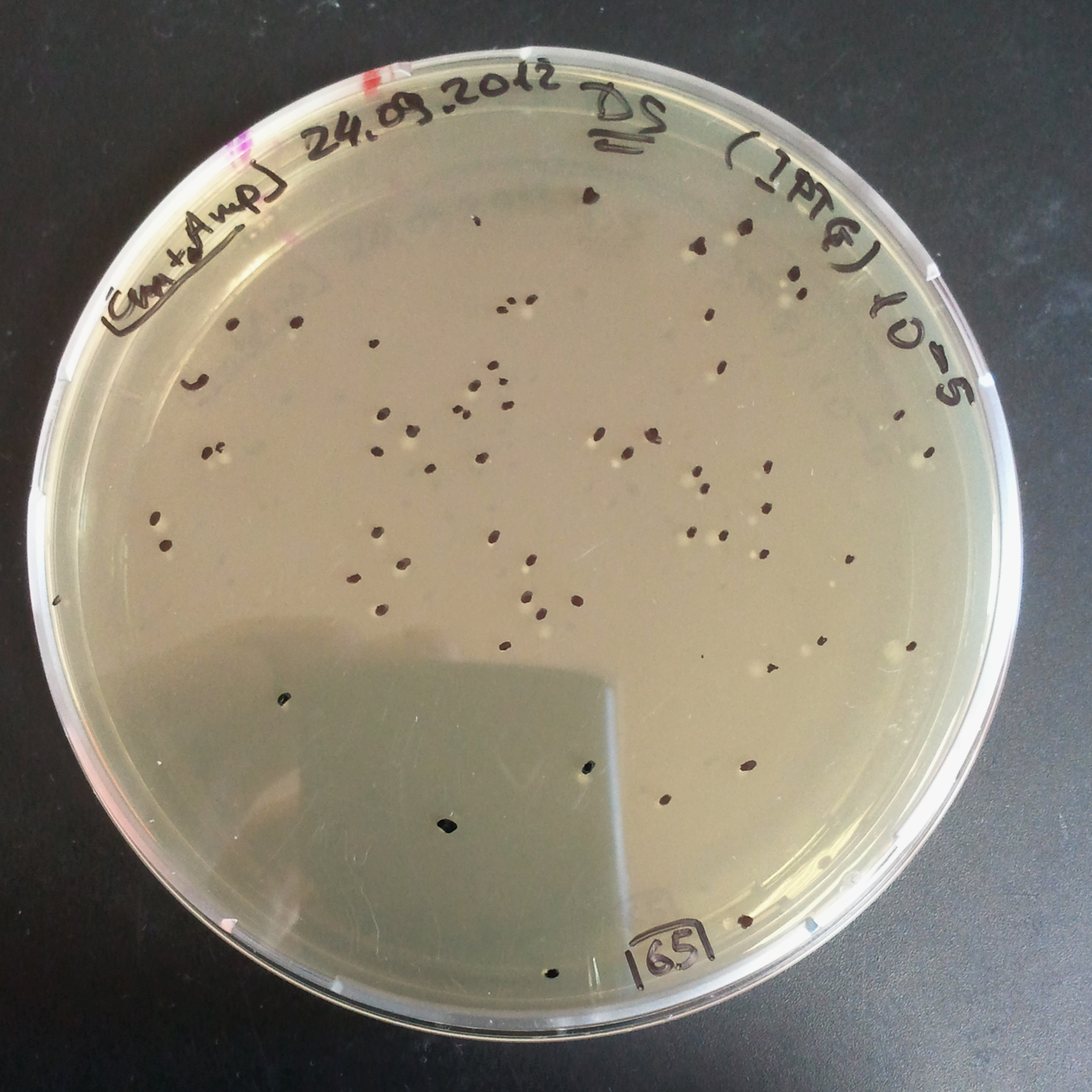
- The second culture with Cm and IPTG.
- Incubate bacteria to have OD = 0.5
- Plate colonies from each tube on two different plates:
- Selection with Cm and Amp.
- Selection with Cm.
The advantage of the new version of our protocol is that we add IPTG at the very low OD, so we expect to have better induction of the meganuclease.
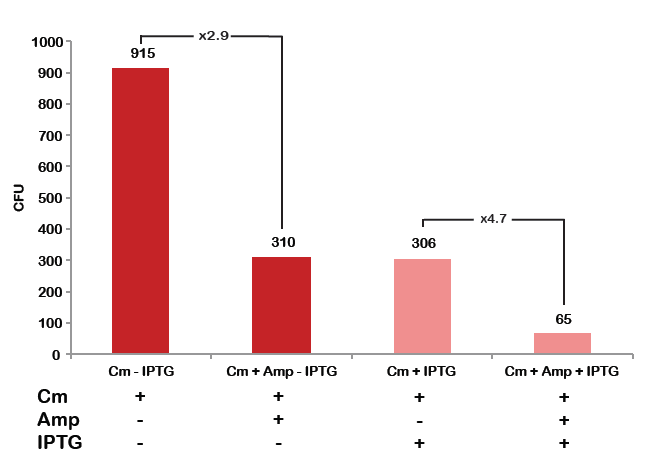
Experimental results:
In much the same way with our previous experiment, we counted and compared the number of colonies on four plates corresponding to 4 conditions:
- First, we compared the number of CFUs formed by the cells where we induced the I-SceI expression, plated with a single antibiotic and with both antibiotics. We expect that there would be much less colonies on the double antibiotic plate, because the Ampicillin carrying plasmid would be lost not only due to the absence of selection during growth and basal expression level of I-SceI (as for the control), but also due to the digestion by the restriction enzyme.
- As control, we compared the number of CFUs formed by non-induced cells, plated with a single antibiotic and with both antibiotics. We expected that the plate with only Cm selection would have the biggest number of colonies. A smaller number would be on the plate with two antibiotics (Amp and Cm). It could be explained by the loss of the plasmid carrying Amp resistance and basal expression level of the I-SceI meganuclease.
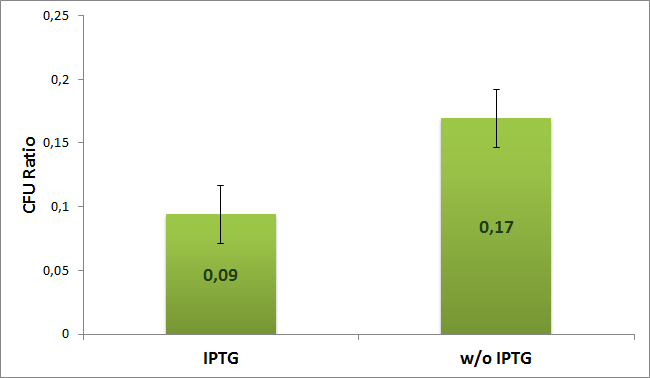
We calcultaed the ratio between plates with single and both antibiotics for both induced (with IPTG) and uninduced (w/o IPTG) case according to the formula:
The experiment was repeated 4 timed. Average value with error bars for both cases are shown on the next diogram:
From the diagram we can see 2 times difference between induced and uninduced case. So results suggest us that the plasmids with AmpR which carry I-SceI restriction site are digested by I-SceI meganuclease.
 "
"