Team:Yale/Project
From 2012.igem.org
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Biological diversity is a very powerful tool, but we are limited by the number of model organisms available to us today. Wouldn't it be wonderful to harness the cryptic pathways of any organism? This year, Yale tried to make a key step in that direction.
Background
The project this year relies on two relatively new technologies, Recombineering and Multiplex Automated Genome Engineering (MAGE).
Recombineering
Recombineering is recombinase mediated genetic engineering. It harnesses the function of "recombinases" like the bacteriophage Lambda protein Beta that bind single stranded pieces of DNA and target them to the lagging strand of the DNA replication fork. Because the replication fork is conserved over all Kingdoms of life, this technique, with the right recombinase, could potentially work in any species.
MAGE
"MAGE" is recombineering on steroids. Instead of introducing one type of synthetic DNA, many areas of the genome are targeted. By repeating the procedure over and over again in discrete cycles, an incredible amount of diversity can be created. This can be used in a wide range of applications, including pathway optimization and entire codon engineering. So far, MAGE has only been developed for E. coli.
But MAGE does have some challenges that limit its efficiency, principally among them the costly procedure of electroporation. Although this is the most efficient way to get the mutagenic oligonucleotides into the E. coli cells, it kills 90% of the cells in solution and requires a recovery growth period. This dramatically lowers efficiency rates.
We aim to increase both the diversity of MAGE-able organisms and the efficiency of the technique by introducing recombinase-mediated genetic engineering to naturally competent organisms.
Project Overview
At the most fundamental level, the successful development of an optimized and fully functional MAGE system in a novel organism requires addressing five points:
- Selection of an industrially-relevant strain of the species of interest
- Optimization of synthetic oligonucleotides for efficient genome modification
- Construction of strains with deficiencies in their mismatch repair (MMR) pathways
- Isolation of ssDNA-binding recombinase proteins that bind mutagenic oligos
- Implementation of the MAGE methodology through continuous cycles of recombination
Keeping those five main criteria of MAGE in mind, we propose to introduce the process in the naturally competent species Bacillus subtilis (Bs) and Acinetobacter baylyi (Ac, or ADP1) in an effort to enhance the efficiency and diversity of this method. Our collective research on each point is presented here, along with some further background where necessary.
Upcoming goals
Our project is multifaceted, but we have already completed most of the component projects and are rapidly approaching a proof of concept, and as of October 3 expect within weeks to have integrated our test cassette onto the genome of a naturally competent cell, transfected it with the beta protein submitted to the Parts Registry, and observed its behavior after exposure to a mutagenic oligo pool. If successful, by November we will have demonstrated an easier and widely applicable method for rapid mutagenesis and directed evolution.
Selection of an industrially-relevant strain of the species of interest
Bacillus subtilis
Our efforts in Bs will be focused on two different MMR-deficient strains obtained separately:
- 1A833 - from BGSC (obtained 6/20 via spore dots on filter disks)
- Reference: BTS13 in Smith et al. (2001)
- Gene descriptor: ΔMutSL::Spc
- Growth conditions: LB, TBAB, Nutrient Agar, or other rich medium containing spectinomycin (100 ug/mL); 37o C
- Ancestry: Strain PY79 (BGSC no. 1A747 - expected arrival imminent)
- Protrophic, shares common ancestor as 168 with 28.6kb W23 island surrounding trpC locus
- PERM739 - from Pedraza-Reyes (obtained 7/12 via agar plates of questionable sterility)
- Reference: Pedraza-Reyes and Yasbin (2004)
- Gene descriptor: ΔMutSL::Neo
- Growth conditions: TBAB, Penassay broth (PAB) for liquid cultures, both supplemented with Neo
- NOTE: Waiting for author response re: use of LB instead of TBAB, and recommended concentration of Neo
- Ancestry: YB955 (hisC952 metB5 leuC427 xin-1 SpβSENS)
- Prophage-"cured" auxotrophic derivative of 168
As both strains are already deficient in mismatch repair, Item 3 (Construction of strains with deficiencies in their mismatch repair pathways) has also been addressed by our choice of strains. The knockout of MutSL has been confirmed in 1A833 by selection with spectinomycin in both the plating of the rehydrated spore disks and the O/N culture for glycerol stocks.
Acinetobacter baylyi
Efforts in Acinetobacter will be focused on one strain.
- ADP1 ΔmutS - from Genescope (obtained 6/21 in liquid culture from Prof. Isaacs)
- Reference: de Berardinis et al., 2008, supplementary table 1 - Media:Msb200810-s2.xls
- Gene Decriptor: ΔmutS::kanr
- Growth Conditions: Same as E. coli, LB media
- Ancestry: Acinetobacter baylyi ADP1
As this strain is already deficient in mismatch repair, Item 3 (Construction of strains with deficiencies in their mismatch repair pathways) has also been addressed by our choice of strains.
Optimization of synthetic oligonucleotides for efficient genome modification
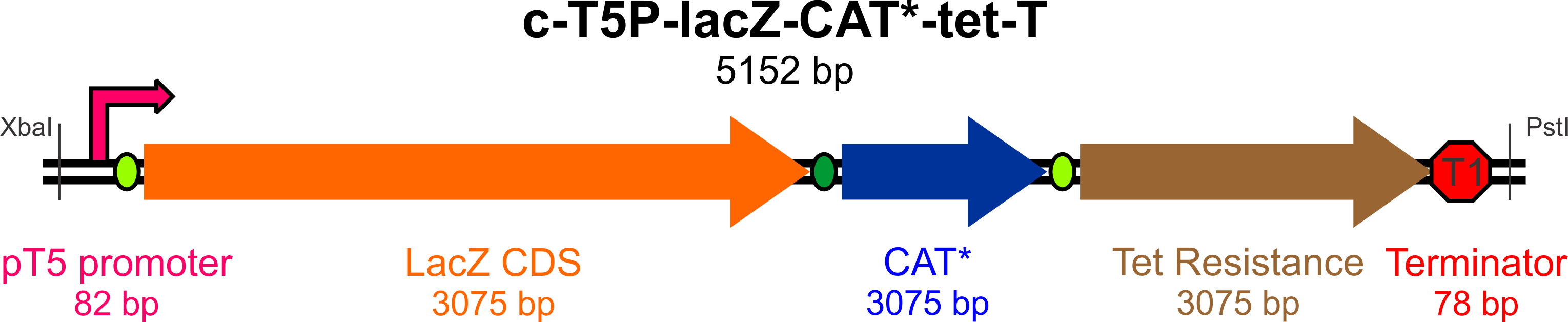
In an effort to streamline the optimization experiments for Bs and Ac, we aim to build a "universal" construct (using E. coli as a cloning behicle) that will allow us to test the success and efficiency of targeted incorporation of mutagenic oligonucleotides at the lagging strand of the replication fork.
This construct will require the following parts to be usable in both Bs and Ac:
- An active lacZ gene, to be inactivated by a mutagenic oligo to yield white colonies on Xgal/IPTG plates
- A promoter for lacZ capable of high expression in both Gram-negative (Ac) and Gram-positive (Bs) bacteria
- A defective copy of a gene conferring antibiotic resistance (plus its native promoter), to be corrected by a mutagenic oligo to yield colonies resistant to the antibiotic of choice (Chloramphenicol)
- An active (unique) antibiotic resistance gene with its own promoter, which will be used to select colonies (for Bs, Ac, and Ec) that have successfully incorporated the construct (we chose Tetracycline)
- An origin site recognized by the replication machinery for all three species
- Transcription terminators flanking the origin of replication to prevent RNA read-through to the plasmid origin part
- Unique restriction sites at appropriate locations for mobility of the construct and some of its elements
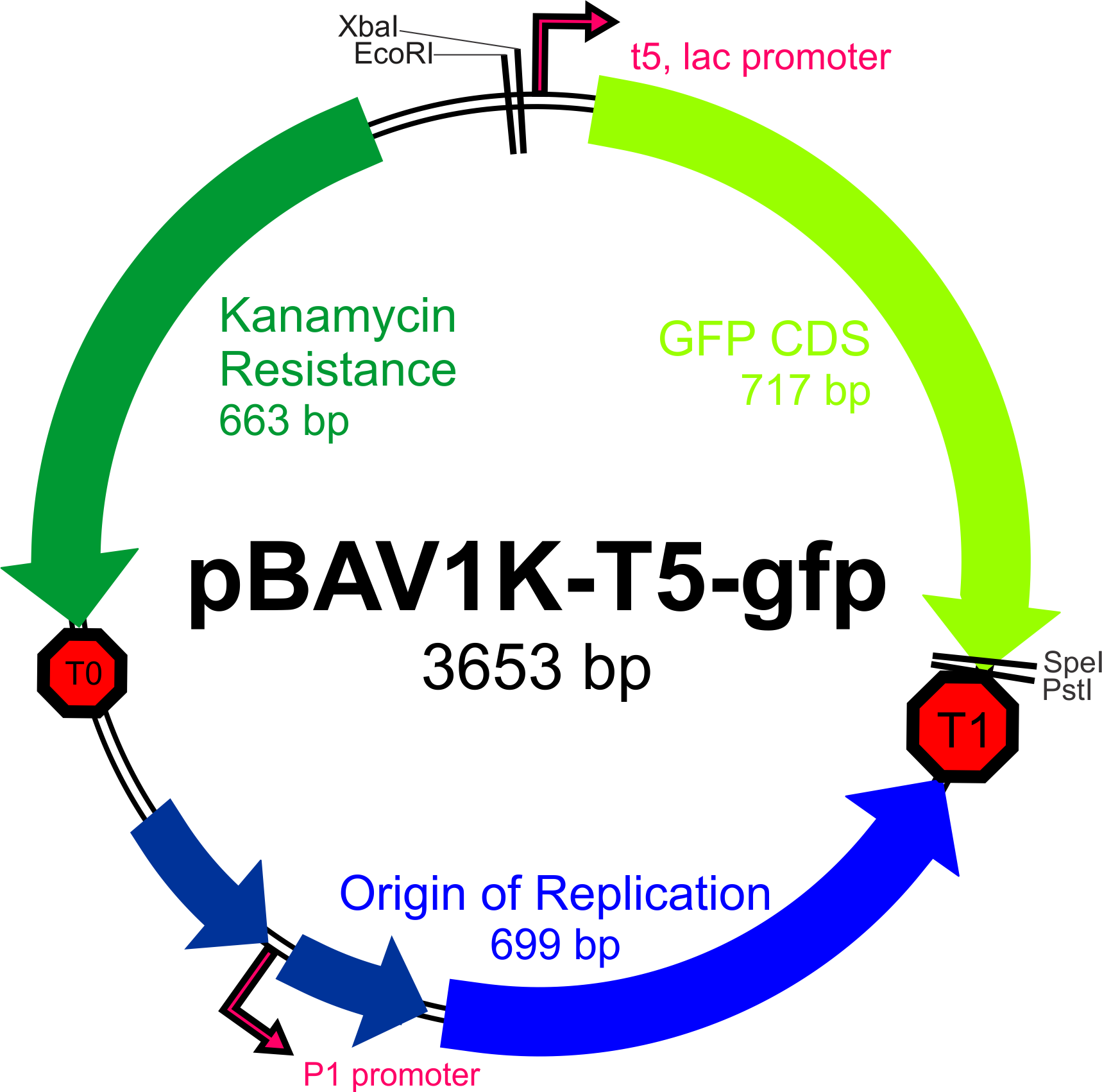
The broad-host range expression vector pBAV1K-T5 (as described by Bryksin and Matsumura, 2010) has been selected as the backbone of our construct, as the authors were able to obtain high copy numbers and high levels of expression in all three of the species we will be working with. Furthermore, this plasmid contains an IPTG-inducible T5 promoter within a BioBrick multiple cloning site, an optimized ori site flanked by the T1 and t0 terminators, and unique restriction sites for the incorporation of selectable markers driven by their own promoters.
Our first priority upon receiving pBAV1K-T5-gfp from Addgene will be to verify that the plasmid can be incorporated (and can replicate) in Bs, Ac, and Ec.
lacZ reporter
Defective selectable marker
Active selectable marker
Miscellaneous plasmid pieces: ori elements, terminators, restriction sites
Construction of strains with deficiencies in their mismatch repair (MMR) pathways
See Item 1 (Choosing an industrially-relevant strain) regarding the MMR-deficient ΔMutSL strains chosen for our experimentation.
Maintenance of bacterial samples
As with the samples we have already received, the strains arriving imminently (Bs: 168 and PY79 for controls, PERM739; Ec: DH5α containing pBAV1K-T5-gfp) will be inoculated in O/N cultures for glycerol stocks.
Verification of the universal replication of pBAV1K-T5-gfp in Ac, Bs, and Ec
Upon receiving pBAV1k-T5-gfp (Bryksin and Matsumura) from Addgene, we will test the unaltered expression vector for transformation in electrocompetent cells of the following strains:
- EcNR2 (ΔmutS, λ red+)
- Bs 1A833 (mutSL::Spcr)
- Bs 1A40 (lys-3 trpC2 metA10)
- ADP1 WT
Transformation of B. subtilis and Acinetobacter baylyi with pBAV1K-based construct
Primer Design
Using Geneious in conjunction with sequences for the various components (obtained from various sources), we designed primers
cT5 (normal)
- Primers have been designed according to overhang specifications from Prof. Isaacs
- Tm overhang: 60-65 oC
- Tm annealing: 55-60 oC
- Low GC count or AT percentage
- Primers have been checked for secondary structures
- Some primers worrisome for %GC
- Primers arrived (07/06/12) from Keck
(Redesigned, see below.)
- Primers for restriction enzyme ends:
- Primers have been checked for secondary structures
| Name | Tm (Tm overhang/Tm anneal) | Length (bp) | Sequence (overCUTSITEanneal) |
|---|---|---|---|
| pAf | 66.64 (25.23/58.79) | 30 (10/20) | 5'-tgacTCTAGAtttgctttgtgagcggataa-3' (XbaI) |
| pAr | 69.08 (30.28/55.61) | 25 (10/15) | 5'-gtaaCTGCAGgagagcgttcaccga-3' (PstI) |
- New Primer List (19Jul12):
| Name | Tm (Tm overhang/Tm anneal) | Length (bp) | Sequence (overhangANNEAL) |
|---|---|---|---|
| pBf | 74.16 (68.90/56.06) | 66 (46/20) | 5'-tgactctagatttgctttgtgagcggataacaattataatagattcAATTGTGAGCGGATAACAAT-3' |
| pBr | 79.55 (75.05/56.42) | 60 (44/16) | 5'-gtaactgcaggagagcgttcaccgacaaacaacagataaaacgaAAGGCCCAGTCTTTCG-3' |
| p2f_2 | 74.20 (69.30/55.65) | 66 (46/20) | 5'-aattgtgagcggataacaattactagagaaagaggagaaatactagATGACCATGATTACGGATTC-3' |
| p2r_2 | 72.60 (63.89/55.41) | 50 (30/20) | 5'-tctccatacctcatcactcctcaatattacTTATTTTTGACACCAGACCA-3' |
| p3f_2 | 70.57 (62.42/56.57) | 54 (32/22) | 5'-caaaaataagtaatattgaggagtgatgaggtATGGAGAAAAAAATCACTGGAT-3' |
| p3r | 72.61 (62.18/58.07) | 50 (37/13) | 5'-ttgttagatttcatatattctttctcctctttcttaaTTACGCCCCGCCC-3' |
| p4f | 71.04 (62.15/56.45) | 50 (30/20) | 5'-ggcgtaattaagaaagaggagaaagaatatATGAAATCTAACAATGCGCT-3' |
| p4r_2 | 81.11 (76.12/57.88) | 59 (44/15) | 5'-aaggcccagtctttcgactgagcctttcgttttatttgatgcctTCAGGTCGAGGTGGC-3' |
Sequencing Primers (ordered 24 Jul 2012)
| Name | Tm | Length (bp) | %GC | Sequence |
|---|---|---|---|---|
| p2f_seq | 61.86 | 27 | 40% | 5'-GCGGATAACAATTACTAGAGAAAGAGG-3' |
| p2r_seq | 59.45 | 21 | 52% | 5'-CTCCATACCTCATCACTCCTC-3' |
| p3f_seq | 56.14 | 22 | 41% | 5'-GTAATATTGAGGAGTGATGAGG-3' |
| p3r_seq | 61.76 | 22 | 50% | 5'-CCTCTTTCTTAATTACGCCCCG-3' |
| p4f_seq | 60.35 | 25 | 40% | 5'-GGCGTAATTAAGAAAGAGGAGAAAG-3' |
| p4r_seq | 64.01 | 27 | 40% | 5'-GACTGAGCCTTTCGTTTTATTTGATGC-3' |
Sequencing Primers II - full cassette sequencing
| Name | Tm | Length (bp) | %GC | Sequence |
|---|---|---|---|---|
| pCas_1F | 58.97 | 22 | 45.5% | 5'-GGCGTATAACATAGTATCGACG-3' |
| pCas_1R | 60.73 | 20 | 50% | 5'-GTTGCACCACAGATGAAACG-3' |
| pCas_2F | 59.53 | 20 | 50% | 5'-GTTTGTTCCCACGGAGAATC-3' |
| pCas_2R | 62.59 | 21 | 52.4% | 5'-CAGGCTTCTGCTTCAATCAGC-3' |
| pCas_3F | 61.85 | 20 | 55% | 5'-CGATGAGCGTGGTGGTTATG-3' |
| pCas_3R | 59.53 | 21 | 47.6% | 5'-GATCGACAGATTTGATCCAGC-3' |
| pCas_4F | 61.22 | 21 | 52.4% | 5'-GATCGTAATCACCCGAGTGTG-3' |
| pCas_4R | 64.40 | 20 | 60% | 5'-CAGAGGCACTTCACCGCTTG-3' |
| pCas_5F | 63.59 | 21 | 52.4% | 5'-GAAGCAAAACACCAGCAGCAG-3' |
| pCas_5R | 64.88 | 21 | 57.1% | 5'-CAGCAAGTGTATCTGCCGTGC-3' |
| pCas_6F | 62.87 | 21 | 52.4% | 5'-CATTGGCGTAAGTGAAGCGAC-3' |
| pCas_6R | 61.44 | 21 | 52.4% | 5'-GACCAACTGGTAATGGTAGCG-3' |
| pCas_7F | 62.49 | 24 | 45.8% | 5'-GAAGAAGGCACATGGCTGAATATC-3' |
| pCas_7R | 60.31 | 22 | 45.5% | 5'-GCGAAGAAGTTGTCCATATTGG-3' |
| pCas_8F | 62.94 | 20 | 55% | 5'-GCAAGATGTGGCGTGTTACG-3' |
| pCas_8R | 60.52 | 20 | 55% | 5'-GGTGATGTCGGCGATATAGG-3' |
| pCas_9F | 63.33 | 23 | 52.2% | 5'-CTTCGCTACTTGGAGCCACTATC-3' |
| pCas_9R | 64.43 | 20 | 60% | 5'-CGAACGCCAGCAAGACGTAG-3' |
| pCas_10F | 62.89 | 22 | 50% | 5'-GCTTGCGGTATTCGGAATCTTG-3' |
| pCas_10R | 60.89 | 20 | 55% | 5'-GGCGGATTTGTCCTACTCAG-3' |
ADP1 Integration Primers
http://www.nature.com/msb/journal/v4/n1/images/msb200810-f1.jpg
Figure from de Berardinis et al 2008 showing locations of primers. Kan cassette in this case will be replaced with either the cT5 cassette (cas) or Zeocin cassette (zeo).
| Name | Tm (Tm overhang/anneal) | Length (bp) | Sequence (overhangANNEAL) |
|---|---|---|---|
| P3 | 57.51 | 22 | 5'-GTTCGTTATTTCCTCACTCATC-3' |
| P4_cas | 75.08 (63.97/61.78) | 47 (25/22) | 5'-cgctcacaaagcaaatctagagtcaATGTTGGTGTTCCAGCTTTACC-3' |
| P4_zeo | 75.10 (62.85/61.78) | 47 (25/22) | 5'-atgccgatgattaattgtcaacaccATGTTGGTGTTCCAGCTTTACC-3' |
| P5_cas | 77.52 (69.08/58.19) | 44 (25/19) | 5'-tcggtgaacgctctcctgcagttacTTACGCTCAAACAGTCGAG-3' |
| P5_zeo | 75.04 (65.49/58.19) | 44 (25/19) | 5'-gctcgaaggctttaatttgcaagctTTACGCTCAAACAGTCGAG-3' |
| P6 | 61.50 | 22 | 5'-ATGATGTTGTTTGGCGGTTTTG-3' |
| Z.ab-F | 61.15 | 23 | 5'-GGTGTTGACAATTAATCATCGGC-3' |
| Z.ab-R | 61.04 | 22 | 5'-AGCTTGCAAATTAAAGCCTTCG-3' |
Z.ab F and R are to amplify the Zeocin cassette.
Potential problems: According to the IDT oligo analyzer, there are some unfortunate secondary structures/self-dimerization potentials for the longer primers, but I am not sure how to avoid this.
Homolog Library Primers
27 total primers:
| Name | Tm (Tm overhang/Tm anneal) | Length (bp) | Sequence (overhangANNEAL) |
|---|---|---|---|
| pT5_F | 76.37 | 92 | 5'-CTAGTCTAGATTTGCTTTGTGAGCGGATAACAATTATAATAGATTCAATTGTGAGCGGATAACAATTACTAGAGAAAGAGGAGAAAGGTACC-3' |
| pT5_R | 76.37 | 92 | 5'-GGTACCTTTCTCCTCTTTCTCTAGTAATTGTTATCCGCTCACAATTGAATCTATTATAATTGTTATCCGCTCACAAAGCAAATCTAGACTAG-3' |
| pUni_F | 66.89 | 32 | 5'-CTAGTCTAGATTTGCTTTGTGAGCGGATAACA-3' |
| pUni_R | 64.52 | 32 | 5'-GGTACCTTTCTCCTCTTTCTCTAGTAATTGTT-3' |
| p40_F | 72.13 (64.83/55.53) | 56 (35/21) | 5'-gataacaattactagagaaagaggagaaaggtaccATGGCAACTAAAAAACAAGAG-3' |
| p40_R | 66.39 (25.28/58.42) | 32 (10/22) | 5'-gcttactagtCTATTCATTTGTTTCCCCTCCT-3' |
| p41_F | 73.76 (64.83/58.35) | 54 (35/19) | 5'-gataacaattactagagaaagaggagaaaggtaccATGGCTGAAAATGCTGTCA-3' |
| p41_R* | 68.49 (25.28/59.97) | 25 (10/15) | 5'-gcttactagtTCATGCGTTGGGCCC-3' |
| p42_F | 72.44 (64.83/55.76) | 55 (35/20) | 5'-gataacaattactagagaaagaggagaaaggtaccATGTCAACTAACGACGAATT-3' |
| p42_R | 66.56 (25.28/59.83) | 33 (10/23) | 5'-gcttactagtTTAGATCATTGACCCTTGAACCT-3' |
| p43_F | 72.34 (64.83/56.43) | 57 (35/22) | 5'-gataacaattactagagaaagaggagaaaggtaccATGGCAAATGAATTAGGAATCT-3' |
| p43_R | 66.53 (25.28/58.73) | 30 (10/20) | 5'-gcttactagtTTAGAATCCCTCCAAAGGCT-3' |
| p44_F | 72.07 (64.83/57.21) | 59 (35/24) | 5'-gataacaattactagagaaagaggagaaaggtaccATGGGGAATGAATTAATAGTAAGC-3' |
| p44_R | 67.14 (25.54/58.91) | 28 (10/18) | 5'-gcAtactagtTCAGAAAGGGTAAGGGCC-3' |
| p45_F | 72.09 (64.83/55.29) | 56 (35/21) | 5'-gataacaattactagagaaagaggagaaaggtaccATGGAAAAACCAAAGCTAATC-3' |
| p45_R | 66.46 (25.28/58.33) | 31 (10/21) | 5'-gcttactagtCTAAGAAGCTAAAGGCTGTGT-3' |
| p46_F | 73.48 (64.83/57.67) | 54 (35/19) | 5'-gataacaattactagagaaagaggagaaaggtaccATGAGCACAGCAGTACAAA-3' |
| p46_R* | 66.50 (25.28/61.33) | 40 (10/30) | 5'-gcttactagtTTATGATGCCTTTTTCCTTAAAAAATCAGA-3' |
| p47_F | 72.72 (64.83/55.62) | 54 (35/19) | 5'-gataacaattactagagaaagaggagaaaggtaccATGGCAACACAAAAAGTTG-3' |
| p47_R* | 66.21 (25.28/60.97) | 40 (10/30) | 5'-gcttactagtTTAAGCCTTATCCTGATTAGTTTCTTTTTC-3' |
| p48_F | 70.19 (64.83/55.02) | 63 (35/28) | 5'-gataacaattactagagaaagaggagaaaggtaccATGACTGAAAATAATAAATTACAAACTA-3' |
| p48_R | 66.05 (25.28/58.60) | 31 (10/21) | 5'-gcttactagtTTAAAATGGCTCTTCTTCGCT-3' |
| p49_F | 73.42 (64.83/57.24) | 54 (35/19) | 5'-gataacaattactagagaaagaggagaaaggtaccATGACTAAGCAACCACCAA-3' |
| p49_R* | 66.40 (25.28/61.07) | 39 (10/29) | 5'-gcttactagtTTATTCCTCTGAATTATCGATTACACTGT-3' |
| p50_F | 73.68 (64.83/56.95) | 52 (35/17) | 5'-gataacaattactagagaaagaggagaaaggtaccATGAGTACTGCACTCGC-3' |
| p50_R | 67.27 (25.28/58.80) | 27 (10/17) | 5'-gcttactagtTCATGCTGCCACCTTCT-3' |
| pExo_R | 66.38 (25.28/57.10) | 39 (10/16) | 5'-gcAtactagtTCATCGCCATTGCTCC-3' |
- *Possible temperature issue
| Name | Tm | Length (bp) | Sequence |
|---|---|---|---|
| pTest_F | 63.88 | 35 | 5'-CGGTGATATTCTCATTTTAGCCATTTATTATTTCC-3' |
| pTest_R | 65.48 | 28 | 5'-GAGCCTTTCGTTTTATTTGATGCCTCTG-3' |
Other Primers
| Name | Tm (Tm anneal) | Length (bp) | Sequence (overhangANNEAL) |
|---|---|---|---|
| Zeo A (F) | 69 (57.13) | 35 (15/20) | 5'-CTATCAACAGGAGTCAGCTTGCAAATTAAAGCCTT-3' |
| Zeo B (R) | 66.26 (61.15) | 38 (15/23) | 5'-ATCCTATATTTAAAAGGTGTTGACAATTAATCATCGGC-3' |
| Zeo A' (R) | 68.38 (56.42) | 35 (15/20) | 5'-TTTAATTTGCAAGCTGACTCCTGTTGATAGATCCA-3' |
| Zeo B' (F) | 63.07 (50.16) | 39 (15/24) | 5'-TTAATTGTCAACACCTTTTAAATATAGGATTTCATTTTC-3' |
| BH mod F | 67.11 | 27 | 5'-gacgtcTTTGCTTTGTGAGCGGATAAC-3' (AatII) |
| BH mod R | 69.62 | 25 | 5'-gacgtcGAGAGCGTTCACCGACAAA-3' (AatII) |
| LacZ KO | - | 90 | 5'-TCAGGATATGTGGCGGATGAGCGGCATTTTCCGTGACGTCTCGTAGCTGCATAAACCGACTACACAAATCAGCGATTTCCATGTTGCCAC-3' |
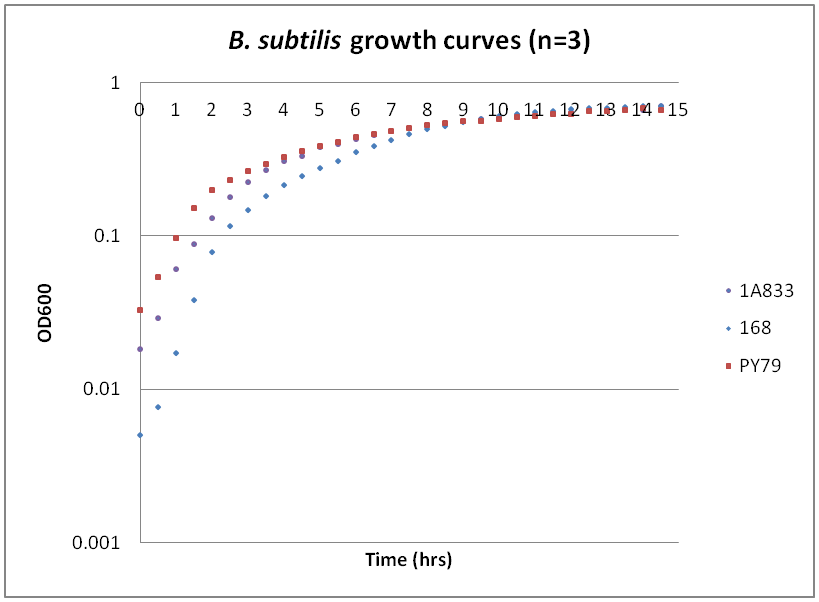
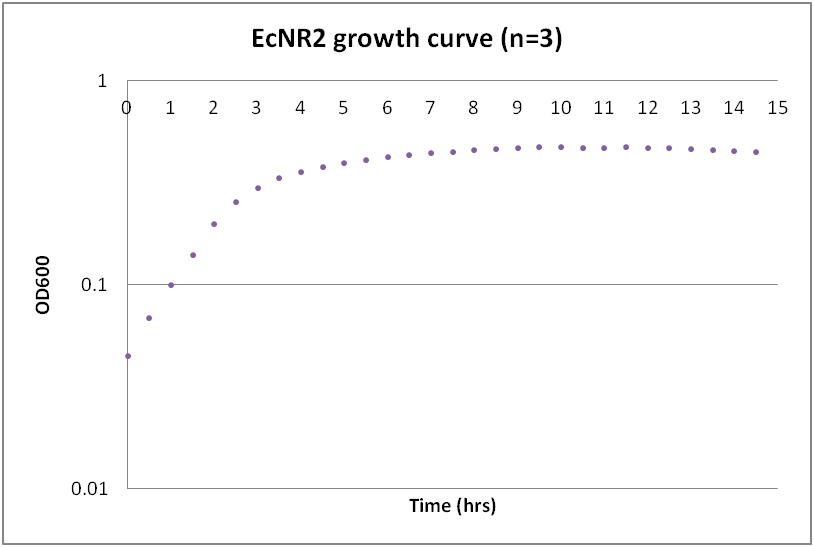
Growth curves for Ac, Bs, and Ec
While troubleshooting the unsuccessful transformation of pBAV1K-T5 in B. subtilis, we thought that it would be beneficial to obtain growth curves for the strains that we were working with. Unlike WT A. baylyi, which is constitutively competent at the stationary phase (12-16 hrs), B. subtilis requires more specific conditions for natural competence, namely starvation and stationary phase growth. Since each of the protocols that we looked at called for slightly different OD600 values – each set of which were largely for the WT strain (168) – we thought that characterizing the growth of all the B. subtilis strains would provide more insight into the optimal conditions for transformation. The growth curves here were generated by growing starter cultures (n=3) of each of the strains in a 24-well plate that was kept incubating in a plate reader at 34⁰C with shaking for 14.5 hours. The OD600 values were recorded automatically every 5 minutes, but the plots show the points at half-hour increments for clarity. What we found was that each of the different strains of B. subtilis showed notable differences in the mid- to late-exponential phase of growth, a fact that can be used in further efforts to transform our cassette-containing vector into B. subtilis via natural competence.
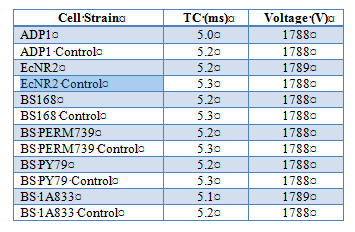
Effect of electroporation on competency in Ab and Bs
To further test competency, we utilized electroporation to make Acinetobacter baylyi and Bacillus subtilis electrocompetent. A.baylyi strain ADP1 (wild type), B. subtilis strains BS168, BS PERM739, BS PY79, BS 1A833, and E.coli strain EcNR2 (E.coli strains implemented as a control due to known success with electroporation). Cells were inoculated from frozen glycerol aliquots and grown O/N in 10mL of LB media until OD600 reading was between 0.4 and 0.8 (midlog phase). Cells were prepared for electroporation by washing cells two times in ice-cold Nuclease-free water and by suspending in Nuclease-free water as the electroporation buffer (exact protocols available {see end of document}). Kanomycin resistance was introduced into the buffer to test successful uptake of plasmids. Electroporation conditions using BioRad Gene Pulser were as follows: 1.8kV, 200Ω, 25uF. Results are as follows:
Generating a beta protein library
We have succeeded in isolating DNA sequences of 16 different lambda red system/beta homolog variants as well as integrating a strong T4 promoter upstream of each. As of right now, at least 6 of them have been integrated into a plasmid compatible with ADP1 and B. subtilis, while identification and confirmation of others is pending.
- To each homolog isolated, we added the T5 promoter (pBAV1K) and XbaI and SpeI sites
- We cloned into pBAV1K and sequenced:
- bet
- recT
- plu2935
- orfC
- s065-exo
- recT-exo
- Other homologs in progress
- We cloned into pBAV1K and sequenced:
- Re-cloned into pSB1C3 (Biobrick)
- bet - BBa_K810000
Helpful Links
On MAGE and its Components
- The original MAGE [http://www.nature.com/nature/journal/v460/n7257/pdf/nature08187.pdf paper] (Wang et al., 2009) with [http://www.nature.com/nature/journal/v460/n7257/extref/nature08187-s1.pdf supplementary information] outlining the optimization of oligos for MAGE in EcNR2 (mutS-, λ-red+).
- Harvard's 2011 iGEM team's simplified outline on MAGE.
- A [http://www.pnas.org/content/105/5/1626.full.pdf?with-ds=yes paper] (Datta et al.) analyzing β-homologs from various Gm- and Gm+ bacteria and their phages for recombinase activity.
On the Strains Chosen for Experimentation
Acinetobacter baylyi ADP1 (formerly BD413 before 1995)
- A broad [http://nar.oxfordjournals.org/content/32/19/5780.full.pdf overview] of ADP1 (Metzgar et al., 2004) presenting it as an ideal model organism for genetic analysis and genome engineering.
- An [http://www.ajol.info/index.php/ajb/article/view/14828 overview] (El Haleem et al., 2004) of the industrial/biotechnical applications of the Acinetobacter genus
- The [http://www.ncbi.nlm.nih.gov/pubmed/15514110 paper] concerning the ADP1 genome (Barbe et al., 2004)
- The associated [http://www.genoscope.cns.fr/cgi-bin/ggb/thesaurus/gbrowse/thesaurus/ baylyi genome browser]
- The [http://aem.asm.org/content/72/1/932 classification] of ADP1 as a new species, A. baylyi (Vaneechoutte et al., 2006)
- A [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2290942/ paper] concerning the construction of a library of single gene mutants in A. baylyi, including ΔmutS (de Berardinis et al., 2008)
Bacillus subtilis
- A [http://bacillus.genome.ad.jp/ genome browser] for B. subtilis
- The [http://www.bgsc.org website] for the Bacillus Genetic Stock Center (BGSC) in Ohio, where we obtained our MMR-deficient strains and their relevant ancestral strains (for controls). In addition to strains of Bs, the BGSC also has integration vectors and information on Bs alleles and their mutant phenotypes.
- A [http://ac.els-cdn.com/S1097276501004026/1-s2.0-S1097276501004026-main.pdf?_tid=5fa060af740627d2245f82e23a94992d&acdnat=1341514852_2c79bf86cc2b32cd987872207fe9e741 paper] (Smith, Grossman, and Walker, 2001) providing background information on the construction of Bs strain 1A833 (mutSL::Spcr).
- A [http://jb.asm.org/content/186/19/6485.full.pdf paper] (Pedraza-Reyes and Yasbin, 2004) providing background information on the construction of Bs strain PERM739 (ΔmutSL::Neo).
- A historical [http://jb.asm.org/content/190/21/6983.full.pdf overview] (Zeigler et al., 2008) of the various laboratory strains of B. subtilis and their genetic and anecdotal backgrounds.
- A protocol from the 2008 Newcastle iGEM team on inducing competence in Bs via starvation media.
- A two-step protocol (Media:Bacillus_subtilis_competent_cell_tf.pdf) based on Harwood et al.'s "Molecular Biological Methods for Bacillus" that allows prior preparation of competent cells stored as a glycerol stock
On Expression/Integration Vectors and their Components
- A [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0013244 paper] (Bryksin and Matsumura, 2010) describing the design of a "universal" plasmid vector capable of replicating efficiently in various Gm- and Gm+ bacteria.
- A [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3255645/pdf/zam280.pdf paper] (Murin et al., 2011) outlining the construction and utility of inducible expression vectors and integration vectors in ADP1
- A [http://www.sciencedirect.com/science/article/pii/104659289290012L paper] (Trumble et al., 1992) describing the design and efficiency of a Ec/Bs shuttle plasmid with a synthetic promoter optimized for both species.
- Addgene's [http://www.addgene.org/26702/ description] on pBAV1K-T5-gfp (universal expression vector)
- Addgene's [http://www.addgene.org/30505/ description] on pIM1463 (ADP1 integration vector)
On Various Aspects of our Oligo-Optimization construct
- A [http://www.sciencedirect.com/science/article/pii/0378111995006524 paper] (Guérout-Fleury et al., 1995) demonstrating the viability of several different antibiotic resistance cassettes in Bs.
On Methods and Technical Aspects of our Project
- [http://arep.med.harvard.edu/kzhang/cgi-bin/myOligoTm.cgi Kun's Oligonucleotide Tm Calculator]
- [http://www.idtdna.com/analyzer/applications/oligoanalyzer/ IDT Oligo Analyzer]
- A [http://www.synbio.org.uk/dna-assembly/guidetogibsonassembly.html website] with links to info on Gibson Assembly
Confirmation of the universality of the pBAV1K-T5 vector
In order to determine whether the pBAV1K-T5 vector (Bryksin and Matsumura, 2010) would serve as an appropriate universal backbone for our cassette, we did a cross-species transformation experiment with the unaltered pBAV1K-T5-gfp plasmid. For E. coli, we used XL1-Blue cells that were made chemically competent, and for the other two species we used naturally competent cells. The success of transformation was gauged by selection on LB-Kan plates, with a concentration of 20 μg/mL for B. subtilis, and 50 μg/mL for Acinetobacter baylyi and E. coli.
What we found was that the XL1-Blue and ADP1 WT cells were consistently transformed (i.e., growth when expected, selective colonies), while the four B. subtilis strains (168, PY79, PERM739, and 1A833) showed no indications of successful transformation despite several different attempts. While it is possible that pBAV1K-T5 simply does not replicate in high enough copy numbers in B. subtilis, it seems more likely that the problem lie somewhere in the transformation protocols. We initially used a two-step transformation protocol from Harwood’s Molecular Biological Methods for Bacillus, and then tried electroporation, then a one-step transformation method also from Harwood’s protocols. The results were consistently poor, with very little reproducibility for any positive results obtained. Upon correspondence with the authors, we learned that they too had trouble yielding consistent results for B. subtilis.
 "
"