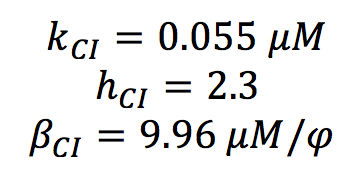Team:Colombia/Modeling/Paramterers
From 2012.igem.org
Team Colombia @ 2012 iGEM
Template:Https://2012.igem.org/User:Tabima
Parameters of the equations
When we want to model a biological process, it is necessary to write the differential equation that model the system which requires a number of constants. If the real value for the constants are unknown, the system can not have any biological sense. These entries are called parameters and they are crucial elements in the mathematical model made in this project.
There are three possible ways to find this parameters:
- i) Literature. There are a lot of studies trying to find biological parameters, such as basal rate of protein production, kinetic constants, Monod's constants, etc. Moreover, there are so many biological systems and only a few of them have been characterized. Thus, it is difficult to find the parameters that are needed.
- ii) Experimental way. If an experiment is made using the biological system of interest, it is possible to find the parameters for the equations that models the whole system. For this project, it was necessary to model the biological system first. Thus, experiments couldn't be performed to find the constant for the differential equations.
- iii) Screening of parameters. Sometimes we don't have the exact number that we need, but we have a rank where it could be or the parameter for a similar biological system, then we can perfom a screening of parameter, where we try to find the value that perfectly fits the reponse of our circuit.
How did we do it?
To find the Pest-busters parameters we developed a standard procedure that can be use any synthetic biology mathematical model. It has four main steps. But before starting to work in it we needed to find as much information in literature as possible.
We found all the CI promoter box parameters, we normalized all the degradation terms with the cell division time and for the other we found acceptable ranges. We used a lot of resources and methods to approximate the limits for this ranges. Here we show an explanation:
a.Basal levels of the proteins:
Determining a rank which this value could be, we described mathematically a very simple process. Suppose there is a usual differential equation:
Where αo is the basal level, γ is the protein parameter for degradation and P is the protein concentration. Thus, we can assume that in steady state conditions we have that dP/dt= 0:
But γ is approximately 1/T, then:
Establishing the rank, we assumed T as φ and we looked for a normal rank of concentration of a protein in a cell [http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=104520&ver=10&trm=protein]. Another important aspect is that there were constitutive proteins and inducible proteins. Thus, we divided the basal levels in two class: i) one for basal proteins and ii) one for constitutive proteins. Finally, following ranges were defined:
Basal proteins range (LuxR, LuxI, HipB and Salicylic Acid):
Constitutive proteins range (HipA7,Sensor, Chitinase, Chitoporin and CBP):
By the other hand, the basal production of HipA has to be constitutive and inducible because the cell has to have three stages. The first stage is always sleep (constitutive) due to HipA effect. Once an impulse of chitin or 3-OH-PAME is presented, it is going to awake the cell due to concentration decrease of HipA . This is the second stage of the cell. The third stage is when cell is slept again which it is achieve through a positive control (inducible). The basal concentration used in the equations was the constitutive one because it is going to have more effect in the response.
b.Protein degradation:
The chosen unit of time was the bacteria life time because it represents when the proteins are diluted and decrease in the cell. We can establish easily the protein degradation using values related to life time of E.Coli. We defined the protein degradation for almost all values as follows:
Again, we found a special case. Degradation of HB is greater, then we assigned it a value of 4/φ.
c. Reaction constant:
This value can vary depending of the described process. Fortunately, we found in literature how to approximate this value [http://www.biokin.com/dynafit/scripting/html/node23.html#SECTION00431000000000000000]. However, we took the approximation and turned it into units described above.
d. Hill coefficients k:
We looked for some reference in different data sources and we found the coefficient of CI ([http://www.sciencemag.org/content/307/5717/1962.short Nitzal et al.2005]). Then, we decided to approximate a rank taking this value as a reference.
e. Hill coefficients n:
We based on the value "n" found for CI( [http://www.sciencemag.org/content/307/5717/1962.short NItzal et al.2005]). In this case, there was an analysis that took in account the number of molecules that interfere with gene activation.
f. Maximum cell production (β):
Considering the concentration of Rubisco, which is about 20 times the average concentration and rate of production of a protein, [http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=107431&ver=1&trm=rubisco], we assign the limits for this range as follows.
CI PARAMETERS
All the parameters for CI activation system were found in the literature. ([http://www.sciencemag.org/content/307/5717/1962.short Nitzal et al.2005]). We only made a change of units.
Once the ranges of the parameters were set, we proceed to do the screening. We tried a lot of techniques and failed, finally we found an acceptable one. Here we present its steps:
- Step 1: Objective function: Define your desired behavior
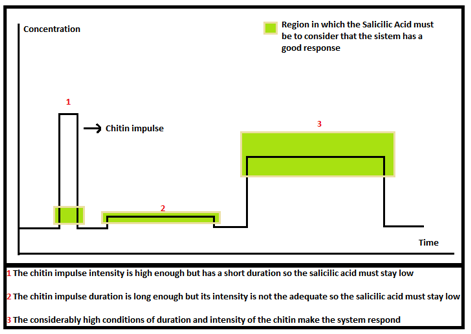
First we define our desired response of the Salicylic Acid. We stablish some ranges where it was suppose to be, depending of the chitin or 3-OH-PAME impulse. We only want our system to respond if the signal is long and intense. Otherwise we don't want our cells to be "awake". The figure below shows this:
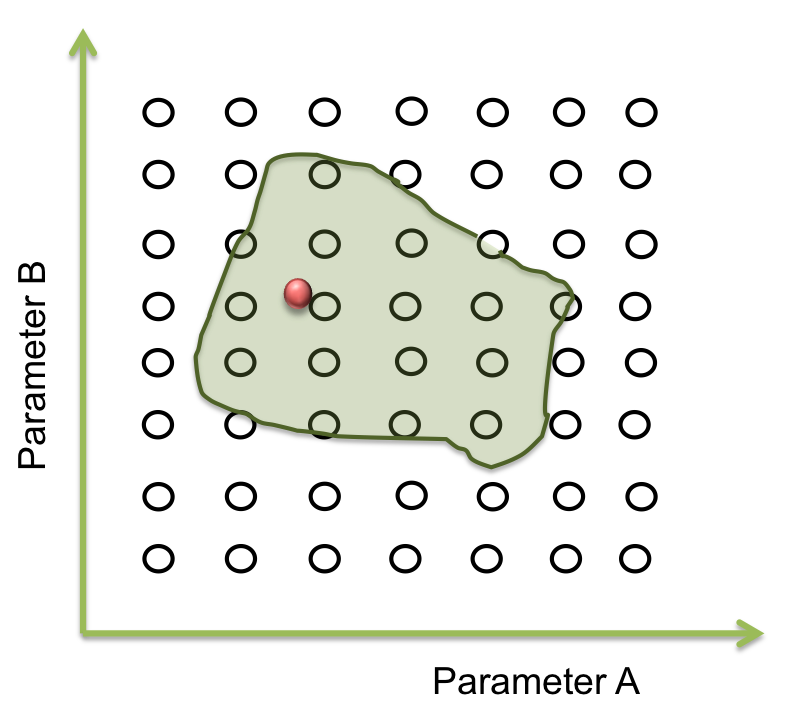
- Step 2. Optimization of parameters: A point within an area
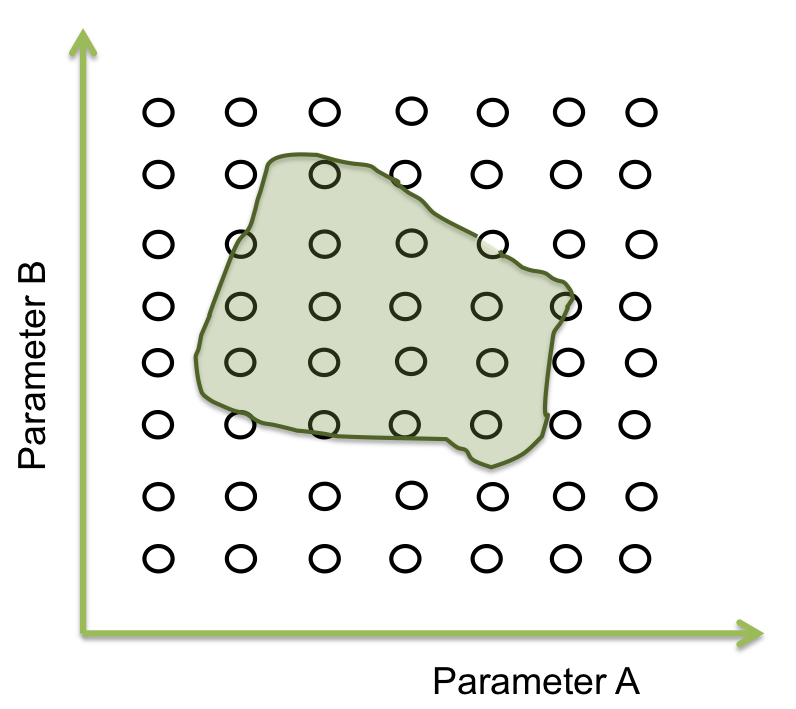
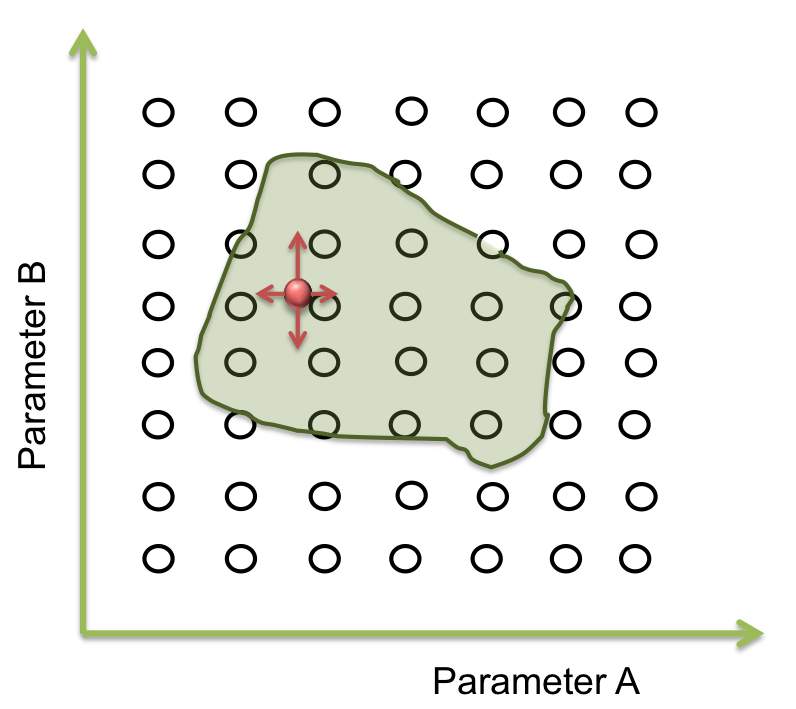
Within the parameter space, there are many sets of parameters that makes the system behaves just like expected; and these can be represented as an area. Our main objective is to find this area.
The goal in this step is to find a point within a specific area and to begin the screening.Suppose we only have two parameters. The plane represents the parameter space and the green area is where the system works.
Now we want a point within these area. It seems easy to identify in two dimensions, but this process can take a lot of time (weeks or even months) when it is a space of more than 10 parameters. So, one way to achieve this process is making an optimization.
A process of optimization takes a function and found maximum or minimum values changing some variable of it. The changing variables are named optimization variables and function is called objective function. If we have some limitations, it is possible to add restrictions to the system, and the function will be optimized without breaking this limitations.
In this case, the objective function is a minimum difference of squares between the points of the expected behavior and the response of the equations with a set of optimization variable. When the distance between these points is minimum, the parameters are found.
This optimization process was done discretizing the differential equations and putting them as restrictions. The same procedure was made for the stable state concentration where the differential equation must be equal to zero (chitin is zero). This algorithm was programmed in the software Gamside.
Unfortunately the firsts results were not satisfactory and a new code it is being developed.
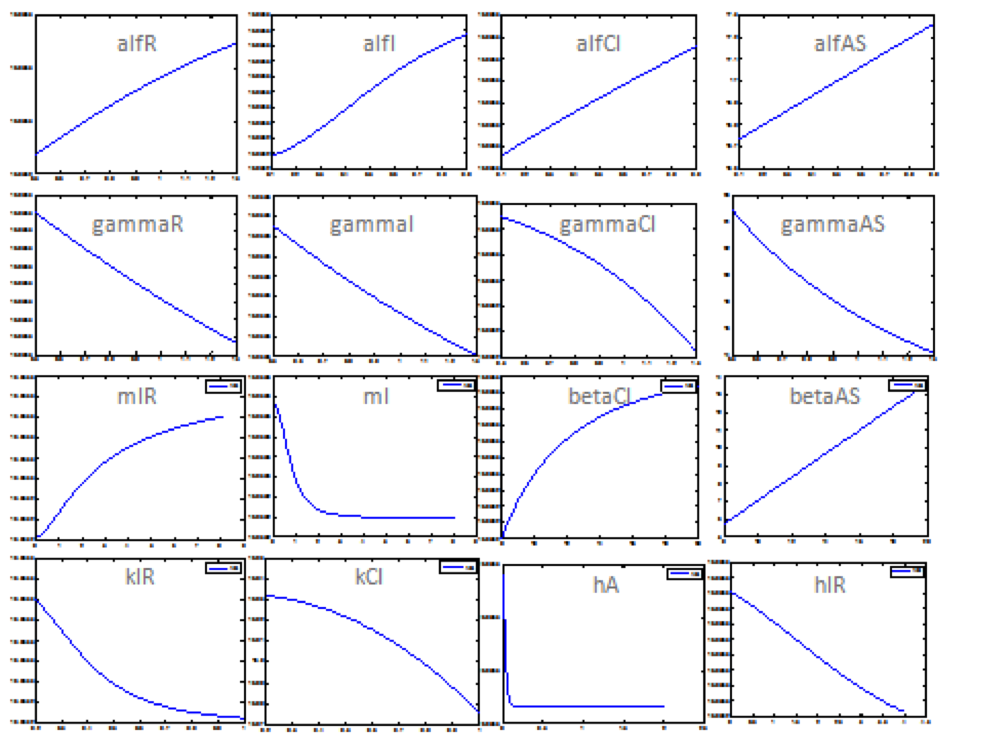
The importance of each parameters in the main outputs behavior (Salicylic Acid, the toxin HipA7 and the antitoxin HipB) was tested. Thus, we did a sensitivity analysis in order to understand how the parameters affect the model.
This test considered the following: establishing the ranges of the parameters (see above), we determined appropriate division for the ranges. Next, we obtained enough data for analysis (according to step size). Then, we iterated each parameter while leaving the others fixed in the MATLAB code.
Note: the exact value for each parameter is not important. It is important their relevance and how they change the response.
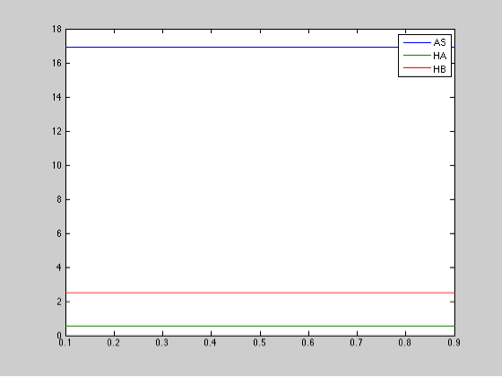
Here is an example:
Parameter: Chitinase production
Range: 0.1-0.9 μM
Step size: 0.01
The diagram above shows that the chitinase production does not have an important effect in salicylic acid, toxin, or antitoxin behavior.
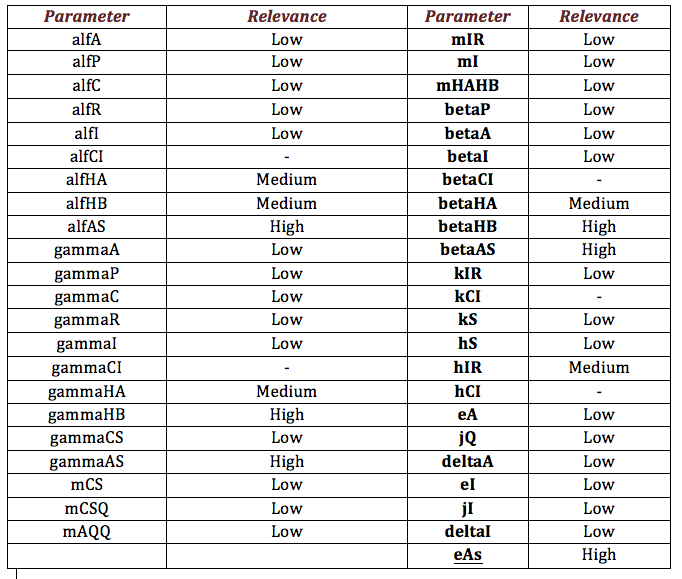
We make this procedure for all the parameters in both systems (Ralstonia and Rust). The results are shown below:
Rust: The following table shows the different parameters involved in the coffee rust detection system and how they affect the final behavior.
The following graphs represent more relevant examples. It shows that rust detection is not directly sensible to some parameters. The blue line is the Salicylic Acid, the green is HipA7 and the red is HipB:
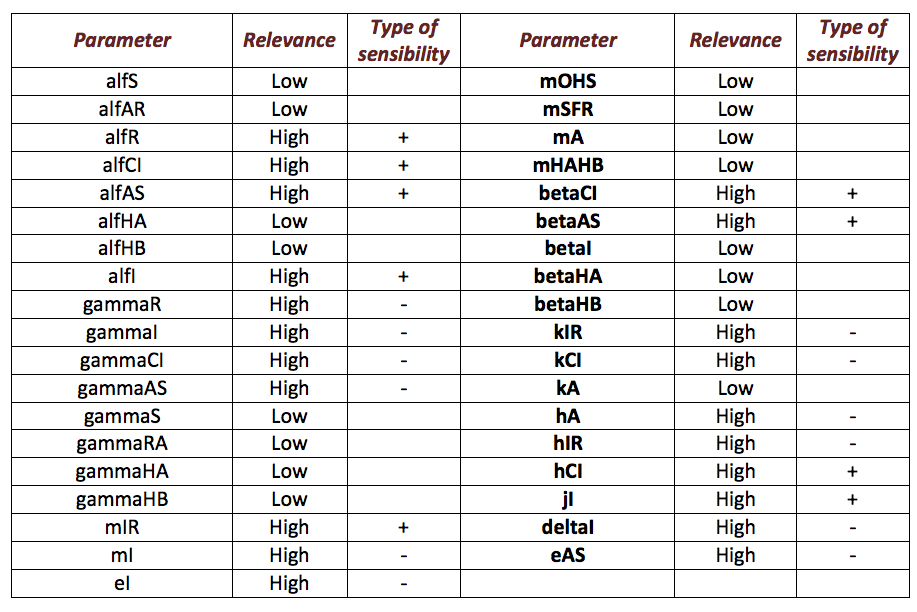
Ralstonia
Below, it is exposed the results obtained from sensibility test for Ralstonia. The next table shows the results obtained. From the results, it is possible extract two important aspects between parameters. The first aspect is identify those parameters that make a relevant change in the model from those that do not. From parameters that are relevant, identify those that are directly or inversely proportional to the salicylic acid response (denoted as “+” and “-“ in the table) is the main objective. This criterion is the second aspect.
The following figure shows the results obtained for almost all the relevant parameters. It sketched the response of the salicylic acid in function of the value for a determined parameter.
Using this results it is possible to know the parameters that affect the response in a major way. Although it is still unknown the exact value of the parameter, it is possible predict how the system is supposed to response. Then, we could proceed to step 2.
- 3.Screening
- If we have a set of parameters, why do we do a screening?
In the last step we found a set of parameters that give the expected response of the system. But this is only a point. What does happen with the rest of points of the area? Does the optimization method “know” if these parameters are real or have biological sense? As long as we specified one single response for the parameters, we don’t know if there are other possible responses of valid solutions. Do they exist?
To answer these questions we performed a screening of the parameters. Beginning at the resulting point of the optimization, we started to move in all directions in a fixed range and then we examined when the acceptable area ends.
How much do we move depends on the sensitivity analysis. If the response is not very sensible to the parameter, we take longer steps than the case which the response is largely sensible.
-Note: Due to long computational time, the code is being parallelized
and we expect to run the tests shortly.
 "
"