Team:UC-Merced/Project
From 2012.igem.org
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Notebook | Safety | Background | Attributions |
|---|
Contents |
Project Summary
The use of microorganisms as a method to obtain hydrogen gas is well documented but there has yet to be a process which can produce the ideal ratio of glucose to hydrogen gas due to a variety of factors. With each method that has been created, only a select number of pathways are modified in order to produce the desired outcome but never close to the ideal 1:4 mole ratio between glucose and hydrogen gas.
With this as the central focus of our project we are planning to remove pathways involved in dark fermentation and adding an entirely new pathway in the hopes of giving our modified E.coli the ability to generate hydrogen gas as a by product. Furthermore, the new pathway can potentially allow better electron transfer efficiency, which can produce the required 1:4 ratio.
Project Inspiration and Idea
Proposed as the ultimate transport fuel, hydrogen holds great promise as an alternative energy source to conventional fossil fuels because it has the potential to eliminate many of the problems that fossil fuels create (Forsberg 2007). Since hydrogen is both a fuel and an energy carrier that can be efficiently converted into other energy carriers, it is especially viable for the fuel cell-based economy in coming years (Jensen et al 2011). However, most hydrogen gas is currently produced using thermochemical reformation of fossil fuels, which results in carbon dioxide waste products (Spormann et al 2005). This major drawback of the current process prevents hydrogen from being considered a truly clean energy source. To avoid the emission of greenhouse gases, the hydrogen source needs to be renewable and carbon-neutral. Biohydrogen production, which is instead catalyzed by microorganisms, presents an attractive and environmentally-friendly conversion of hydrogen energy for the future (Lee et al 2011).
While biohydrogen production is still in its early stage of development, a variety of laboratory- and pilot-scale systems have been developed with promising potential (Lee et al 2011). Several have examined light-driven processes, yet encountered restrictions due to the nature of photobiolysis (Hallenbeck & Benemann 2002). Thus, one emerging area of focus involves taking advantage of the dark fermentation process in microorganisms.
Some systems focused on dark fermentation have investigated the modification of environmental conditions such as pH and temperature or produced recombinant bacteria strains through genetic mutations in order to optimize fermentative capabilities (Valdez-Vasquez & Poggi-Varaldo 2009; Toshinari et al 2007, 2008). For example, Toshinari et al enhanced hydrogen production from Escherichia coli using multiple recombinant strains to introduce multiple mutations into the bacterium (Toshinari et al 2007). Another study by Do et al engineered E. coli for fermentative hydrogen gas production by incorporating NADH-ferredoxin oxidoreductase from the hydrogenosome of anaerobic protozoa (Do et al 2009). At present, however, documented processes have yet to experimentally yield the theoretical ratio of 4 mol hydrogen gas per mol glucose (Hallenbeck & Benemann 2002, Lee et al 2011). Possible factors inhibiting optimum hydrogen gas yield include H2 loss by methanogens in non-sterile systems, limitations of hydrogenase activity, and organic acid concentration among other environmental conditions (Valdez-Vasquez & Poggi-Varaldo 2009). Therefore, strategies to avoid hydrogen gas production inhibition are recommended.
Our team recognized that key elements from several existing systems can be consolidated and modified into a new pathway that may potentially optimize bacterial production of hydrogen gas toward the ideal ratio. Our project focuses on a strategy of producing hydrogen gas from Escherichia coli by simply knocking out and inserting select components to maximize hydrogen production through the dark fermentation process.
Strategy of Utilizing Fermentative Capabilities in E. coli to Produce Hydrogen Gas
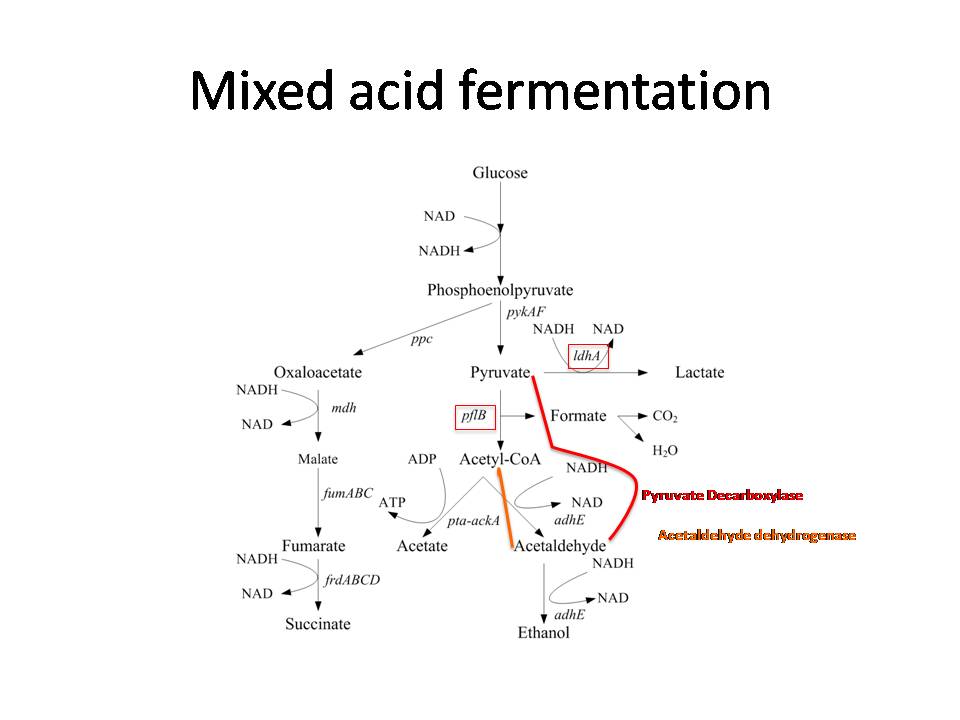
Generally, the mixed acid fermentation of E. coli, which begins at the end of glycolysis, involves the production of lactate, acetate, formate, and ethanol from pyruvate (Cortassa et al 2002).
Furthermore, pyruvate is also involved in the initial steps of dark fermentation, which is not naturally present in E. coli. Therefore, in order to preserve pyruvate for this process, the proteins responsible for producing lactate, acetyl-CoA, and formate were knocked out. The eliminated enzymes were lactate dehydrogenase A (ldhA) and pyruvate formate lyase B (pflB). Concurrently, pyruvate decarboxylase was inserted into the bacteria in order to produce acetaldehyde from pyruvate.
Originally, acetaldehyde in E. coli is converted into ethanol by alcohol dehydrogenase E (adhE) (Gupta et al 2000). Thus, to preserve acetaldehyde for dark fermentation, adhE was also knocked out. Acetaldehyde dehydrogenase was then inserted into the E. coli to convert acetaldehyde into acetyl-CoA. The process of converting acetaldehyde into acetyl-CoA produces 2 NADH and 2 hydrogen protons. In addition, the glycolytic pathway also produces 2 NADH and 2 hydrogen protons. Thus, an expected total 4 NADH and 4 hydrogen protons are produced from one mol of glucose.
NADH is used to convert ferredoxin from its oxidized state into its reduced state (Do et al 2009). This reaction is carried out by ferredoxin oxidoreductase, which is inserted into the bacteria. Ferredoxin then reduces hydrogen protons into hydrogen gas with hydrogenase. As a result of the process, ferredoxin is returned to its oxidized state, enabling the cycle to continue.
We expect that eliminating ldhA, pflB, and adhE from the E. coli fermentation pathway and inserting mhpF, pyruvate decarboxylase, and ferredoxin oxidoreductase will ultimately yield 4 mol of hydrogen gas for every one mol of glucose consumed by the bacteria, thus achieving a more direct line towards hydrogen production.
References:
1. Cortassa S et al. (2002) An Introduction to Metabolic and Cellular Engineering. Singapore: World Scientific: 1-34.
2. Do PM et al. (2009) Engineering Escherichia coli for Fermentative Dihydrogen Production: Potential Role of NADH-Ferredoxin Oxidoreductase from the Hydrogenosome of Anaerobic Protozoa. Appl Biochem Biotechnol 153: 21-23.
3. Gupta S et al. (2000) Acetaldehyde dehydrogenase activity of the AdhE protein of Escherichia coli is inhibited by intermediates in ubiquinone synthesis. FEMS Microbiology Letters 182: 51-55.
4. Hallenbeck PC and Benemann JR. (2002) Biological hydrogen production; fundamentals and limiting processes. International Journal of Hydrogen Energy 27: 1185-1193.
5. Forsberg CW. (2007) Future hydrogen markets for large-scale hydrogen production systems. Int. J. Hydrogen Energy 32: 431–439.
6. Lee D et al. (2011) Dark fermentation on hydrogen production: pure culture. Bioresource Technology 102: 8393-8402.
7. Jensen J et al. (2011) Hydrogen Implementing Agreement: Hydrogen. International Energy Agency, IEA CERT Workshop: 1-23.
8. Spormann AM et al. (2005) Metabolic Engineering of Hydrogen Production in Cyanobacterial Heterocysts. Stanford: GCEP Technical Report: 1-2.
9.Toshinari M et al. (2007) Enhanced hydrogen production from glucose by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol 77: 879-890.
10. Toshinari M et al. (2008) Metabolic engineering to enhance bacterial hydrogen production. Microbial Biotechnology 1(1): 30-39.
11. Valdez-Vasquez and Poggi-Varaldo. (2009) Hydrogen production by fermentative consortia. Renewable and Sustainable Energy Reviews 13: 1000-1013.
Protocols
(click hyperlinks to find protocol)
- [http://www.promega.com/~/media/files/resources/protocols/technical%20bulletins/101/magazorb%20dna%20mini-prep%20kit%20protocol.pdf?la=en Mini Prep] genes of interest (Acetaldehyde dehydrogenase and Ferredoxin oxidoreductase) from respective organisms.
- Nanodrop DNA
- Run Agarose Gel to ensure proper gene fragments are obtained.
- PCR gene fragments to amplify.
- Nanodrop solution
- Extract (Protocol A)
- Digest gene fragments with [http://www.neb.com/nebecomm/products/productr0101.asp EcoRI] and [http://www.neb.com/nebecomm/products/productr0140.asp PstI].
- Ligate into biobrick plasmid pSB1C3.
- Make FMJ39 into competent cells
- [http://tools.invitrogen.com/content/sfs/manuals/oneshottop10_man.pdf Transform] competent FMJ39 cells with pSB1C3+gene.
- Test cells for transformation and hydrogen production by growing in anaerobic conditions.
B.)[http://www.neb.com/nebecomm/ManualFiles/manualE2611.pdf Gibson Assembly Master Mix]
C.)P1 Transduction - Ausubel, F. M. (2001). Current protocols in molecular biology. New York: J. Wiley
D.)Plasmid Transformation [ie. CaCl2 transformation] - Ausubel, F. M. (2001). Current protocols in molecular biology. New York: J. Wiley
E.)LB Agar:Microbiology Laboratory ManualDr. Erin R. Sanders-Lorenz, EditorMicrobiology, Immunology, & Molecular GeneticsUniversity of California – Los Angeles (aka BIO 120L Manual)
F.) The PCR protocol is in the Notebook and was produced by Prof. Garcia-Ojeda.
Future Work
In our current project, we used NADH ferredoxin oxidoreductase as an electron carrier to shuttle electrons to the hydrogenase and lead to hydrogen production. However, there are other pathways which may yield more hydrogen. In future experiments, we would like to do a qualitative analysis on our system and deduce its efficiency. The bacteria transformed for this project (the FMJ39 strain) may have some pathways which may breakdown hydrogen thus reducing yield. We would like to further study the genome of our microbe and minimize the use of such pathways to maximize our yield.
Our transformed bacteria rely on dark-fermentation to produce hydrogen. However, according to Nath, Kumar, and Das, photo-fermentation is a slightly more advantageous pathway because it is capable of harnessing light energy to drive the reactions in the cell (2005). It also has the advantage of breaking down small organic acids, such as acetyl-CoA in our model, and producing more hydrogen. An efficient system may result from the combination of our model and a photo-fermentation model. Our model is capable of using many compounds and shuttle them to hydrogen production but produces byproducts which cannot be broken down further. The photo-fermentation pathway is capable of using light energy to break down these smaller byproducts and produce more hydrogen.
Our current project took a commonly found fermentation process and modified it to create a funnel for hydrogen production. However, to build a complete and reliable system, we would like to add another system which is capable of producing the substrates needed for the current project. We have identified the breakdown of cellulose, common plant matter, as a potential candidate for this.
Combining a system for cellulose breakdown, dark-fermentation hydrogen production, and photo-fermentation hydrogen production will yield a complete system capable of using raw plant matter to produce hydrogen.
References:
[http://www.springerlink.com/content/pam72lj4jur3vkq9/ Nath, Kaushik, Anish Kumar, and Debabrata Das. (2005) "Hydrogen Production by Rhodobacter Sphaeroides Strain O.u.001 Using Spent Media of Enterobacter Cloacae Strain Dm11." Applied Microbiology and Biotechnology. 68.4: 533-541. Print.]
 "
"