Team:NYU Gallatin/Modeling
From 2012.igem.org
Revision as of 03:01, 4 October 2012 by Sararobertson (Talk | contribs)
NYU Gallatin 2012 iGEM Team
Main menu
- HomeHome of Aseatobacter.Home
- TeamThe brains of the operation.Team
- ProjectLearn more about our project.Project
- PartsOur work with the parts registry.Parts
- ModelingHow we put it all together.Modeling
- NotebookLab notebooks, news, and photos.Notebook
- SafetyOur commitment to safety.Safety
- AttributionsGive credit where credit is due.Attributions
- ProfileOfficial iGEM 2012 profile.Profile
Model It
Modeling
Bio Synthetic Pathways

Codonizer
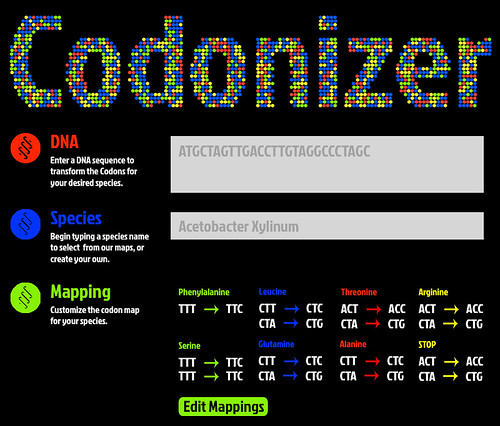
To ensure that our modeling worked, we ran our sequences through a custom developed codon optimizer. Try it our yourself below! Drop in a chunk of DNA data and click "Optimize Codons" to have your sequence optimized for the acetobacter preferred codons.
Upon completion of the iGEM competition we would like to expand our Codonizer software to support other strains. A mockup of what this tools will look like is provided below.
Retrieved from "http://2012.igem.org/Team:NYU_Gallatin/Modeling"
 "
"
