Team:Calgary/Project/HumanPractices/Killswitch/Regulation
From 2012.igem.org


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Regulation/Expression Platform

Tight Regulation
Inducible kill systems are not new to iGEM. Looking through the registry, there are several constructs such as the inducible BamHI system contributed by Berkley in 2007 (BBa_I716462) and tested by Lethbridge in 2011. This uses a BamHI gene downsteam of an arabinose-inducible promoter. Another example is an IPTG inducible Colicin construct (BBa_K117009) submitted by NTU-Singapore in 2008. One major problem with these systems however is a lack of tight control. As was demonstrated by the Lethbridge team, this part has leaky expression when inducer compound is not present. The frequently used lacI promoter has similar problems when not used in conjunction with strong plasmid-mediated expression of lacI. This can be seen in our electrochemical characterization of the uidA hydrolase enzyme (BBa_K902002) shown here. Tight control is not only a problem for kill switch application, but for any application requiring strict regulation. As such, we decided that expanding the registry repertoire of control elements would be useful for our system as well as a variety of other applications.
Introducing the Riboswitch
Riboswitches are small pieces of mRNA which bind small molecules to modify translation of downstream genes. These sites are engineered into circuits by replacing traditional ribosome binding sites with riboswitches. The riboswitch is able to form mRNA conformations around its respective ligand to inhibit or promote binding of translational machinery. Riboswitches can be used in tandem with an appropriate promoter to enable tighter control of gene expression. Given this opportunity for control, and that ligands for riboswitches are often inexpensive small ions, these methods might be a feasible solution for controlling the kill switch in our industrial bioreactor.
We explored 3 different riboswitches, each responsive to a different metabolite (magnesium, manganese or molybdate) that would be cheap to implement into a bioreactor environment. Additionally, we also investigated a repressible and inducible promoter responsive to glucose and rhamnose respectively.
Magnesium riboswitch
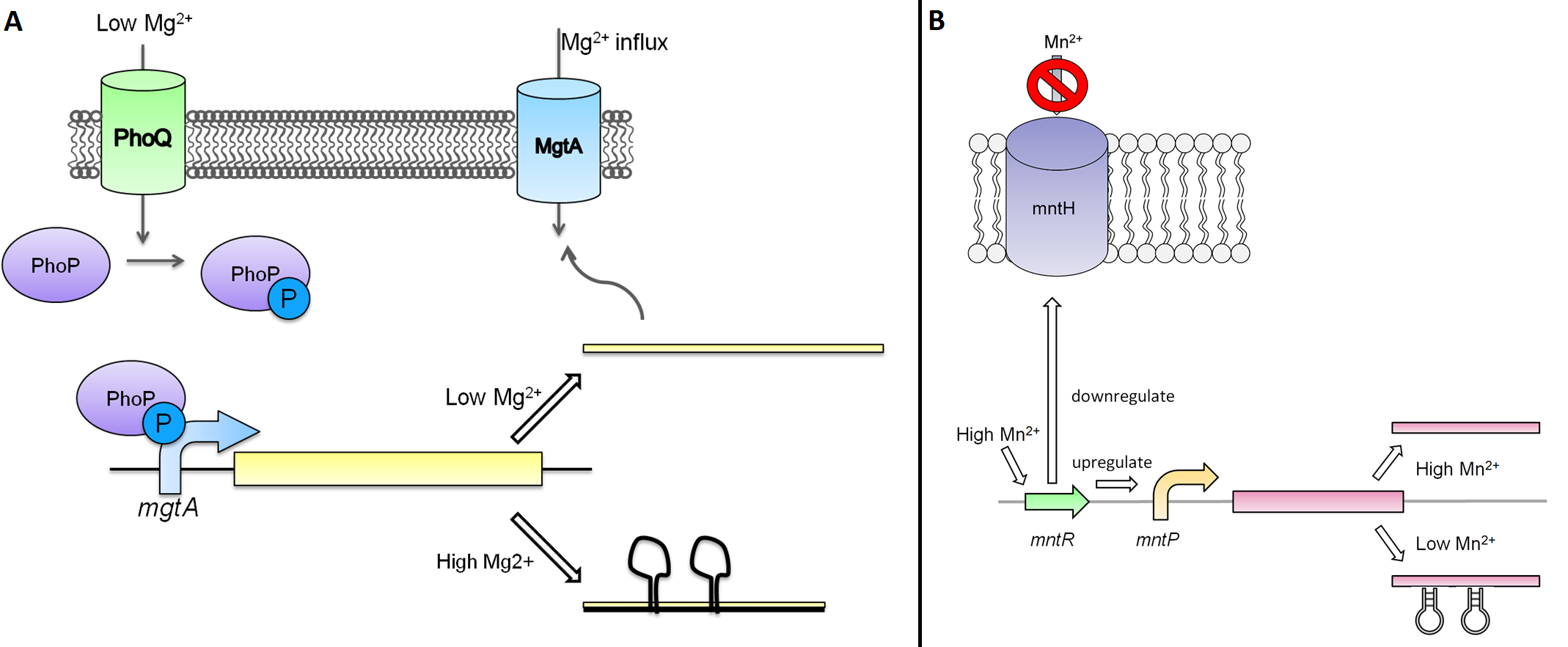
The magnesium riboswitch that we looked at is repressed in the presence of magnesium ions. This system has two control components – a promoter and a riboswitch. Normally the magnesium promoter (mgtA promoter) and the magnesium riboswitch (mgtA riboswitch) are activated if there is a deficiency of magnesium in the cell. The lack of magnesium activates other genes in E. coli to allow influx of magnesium into the cell. The two proteins in the cascade that activate the system are PhoP (BBa_K902010) and PhoQ (BBa_K902011). PhoQ is the trans-membrane protein which gets activated in the absence of magnesium and phosphorylates PhoP. PhoP in turn binds to the mgtA promoter and transcribes genes downstream.
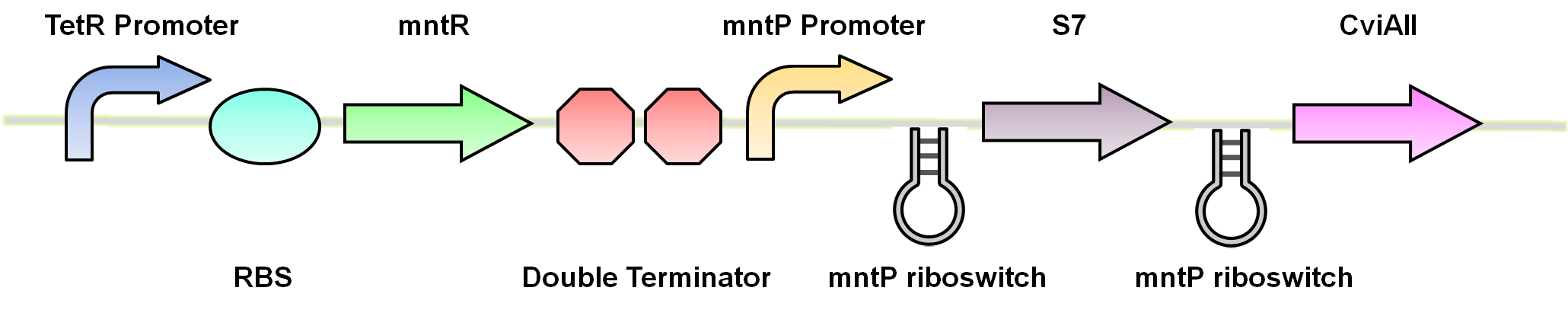
Manganese riboswitch
Manganese is an essential micronutrient. It is an important co-factor for enzymes and it also reduces oxidative stress in the cell (Waters et al, 2011). Despite being an important micronutrient, it is toxic to cells at high levels. MntR detects the level of manganese in the cell and acts as a transcription factor to control the expression of manganese transporter such as mntH, mntP and mntABCDE. In order to regulate these genes mntR (BBa_K902030) binds to the mntP promoter (mntP promoter). The manganese homeostasis is also controlled by the manganese riboswitch mntPrb (BBa_K90274)
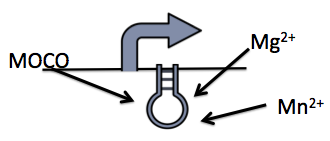
The Moco Riboswitch
The molybdenum cofactor (moco) riboswitch (BBa_K902023) is an RNA element which responds to the presence of the metabolite molybdenum cofactor(MOCO) (Regulski et al, 2008). This RNA element is located in the E.coli genome just upstream of the moaABCDE operon (BBa_K902024), which contain the important moco synthesis genes. Moco is an important co-factor in many different enzymes. The moco riboswitch has 2 regions: an aptamer domain and the expression platform. When moco is present in the cell it will bind to the aptamer region in the riboswitch causing an allosteric change. This allosteric change affects the expression platform by physically hiding the ribosome binding site which prevents translation.
Building the Systems
Using these riboswitches, we wanted to design a system where we would place our kill genes downstream, and then supplement our bioreactor with the appropriate ions to keep the systems turned off. We biobricked and submitted DNA for the the mgtaP (BBa_K902009) and mntP promoter (BBa_K902073) as well as their respective riboswitches (BBa_K902008) (BBa_K902074) and the moco riboswitch (BBa_K902023). In addition, we also biobricked some of the regulatory proteins: PhoP (BBa_K902010), PhoQ (BBa_K902011), mntR (BBa_K902030) and the Moa Operon (BBa_K902024) . Our final system would inovolve constitutive expression of these necessary regulatory elements upstream of our riboswitches and kill genes. An example of the manganese system is shown in figure 3.
Characterizing the riboswitches
GFP testing
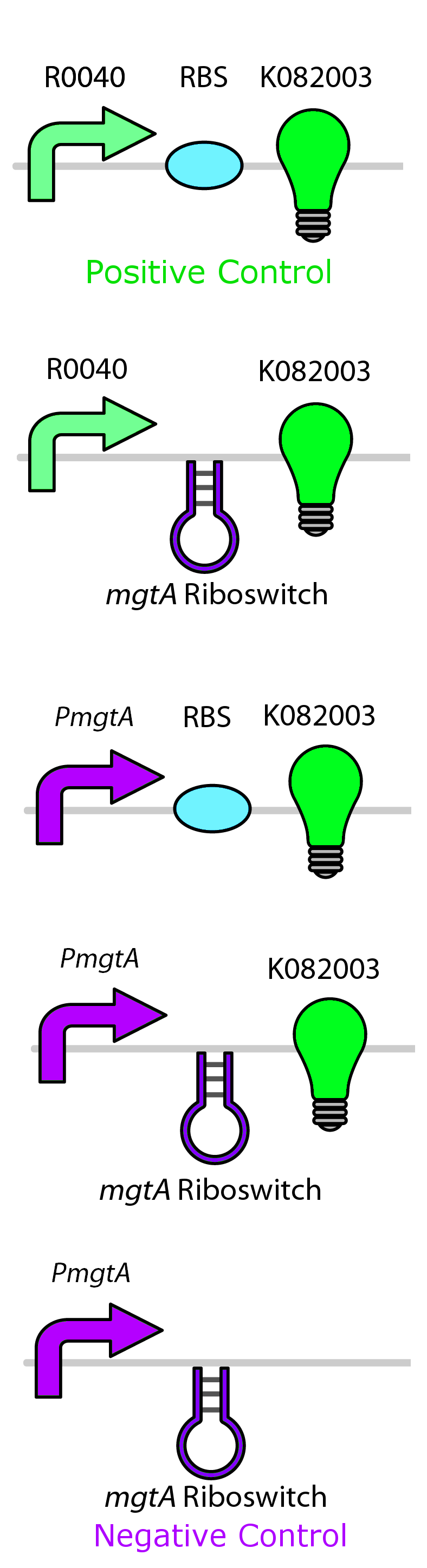
In order to test the control of these promoters and riboswitches, we constructed them independently and together upstream of GFP (BBa_K082003) with an LVA tag. Figure 4 shows these circuits for the mgtA system. Identical circuits were designed for all three systems, however only the top two were needed for the mocoriboswitch system.
We then tested the aforementioned circuits by growing cells containing our circuits with varying concentrations of their respective ions. Our detailed protocols can be found here. We then measured fluorescent output, normalizing to a negative control not expressing GFP.
Results
So far, we have been able to obtain results for our magneisum system, as can be seen in figure 5.
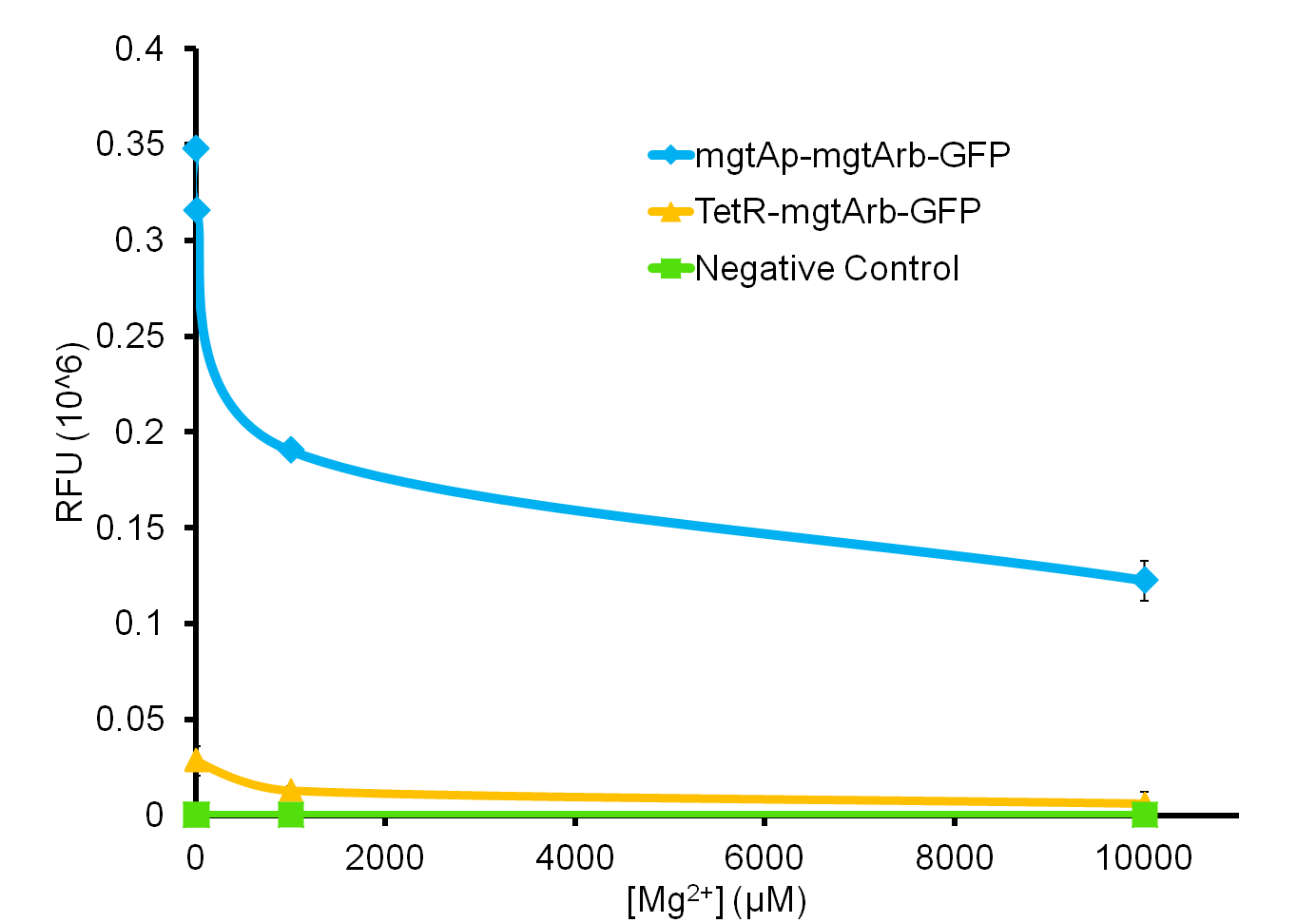
As the graph shows, there is a much larger decrease in the GFP output when the mgtA promoter and riboswitch are working together as compared to the mgtA riboswitch alone under the control of TetR promoter (BBa_J13002). This suggests that having both the promoter and the riboswitch together provides a tighter control over the genes expressed downstream. This also suggests that magnesium riboswitch alone is sufficient in reducing gene expression downstream of a constitutive promoter.
It is important to consider however that the control elements of the system namely PhoP and PhoQ were not present in the circuits tested and therefore there is GFP expression in at the inhibitory concentration (10mM MgCl2). We believe that having the regulatory elements would give us better control and get rid of the leakiness.
Although the magnesium system is highly regulated, it is not a suitable system for the purposes of our bioreactor. The tailings are composed of very high concentration of magnesium- up to 120mM(Kim et al, 2011). As can be seen, this would inhibit the system. Therefore, if our bacteria escapes into the tailings, the kill genes would not be activated and the bacteria would be able to survive. We feel howeever that this could still be an incredibly useful system for other teams for both killswtitch and non-killswitch-related applications, making it still a valuable contribution to the registry.
Kill Gene Testing
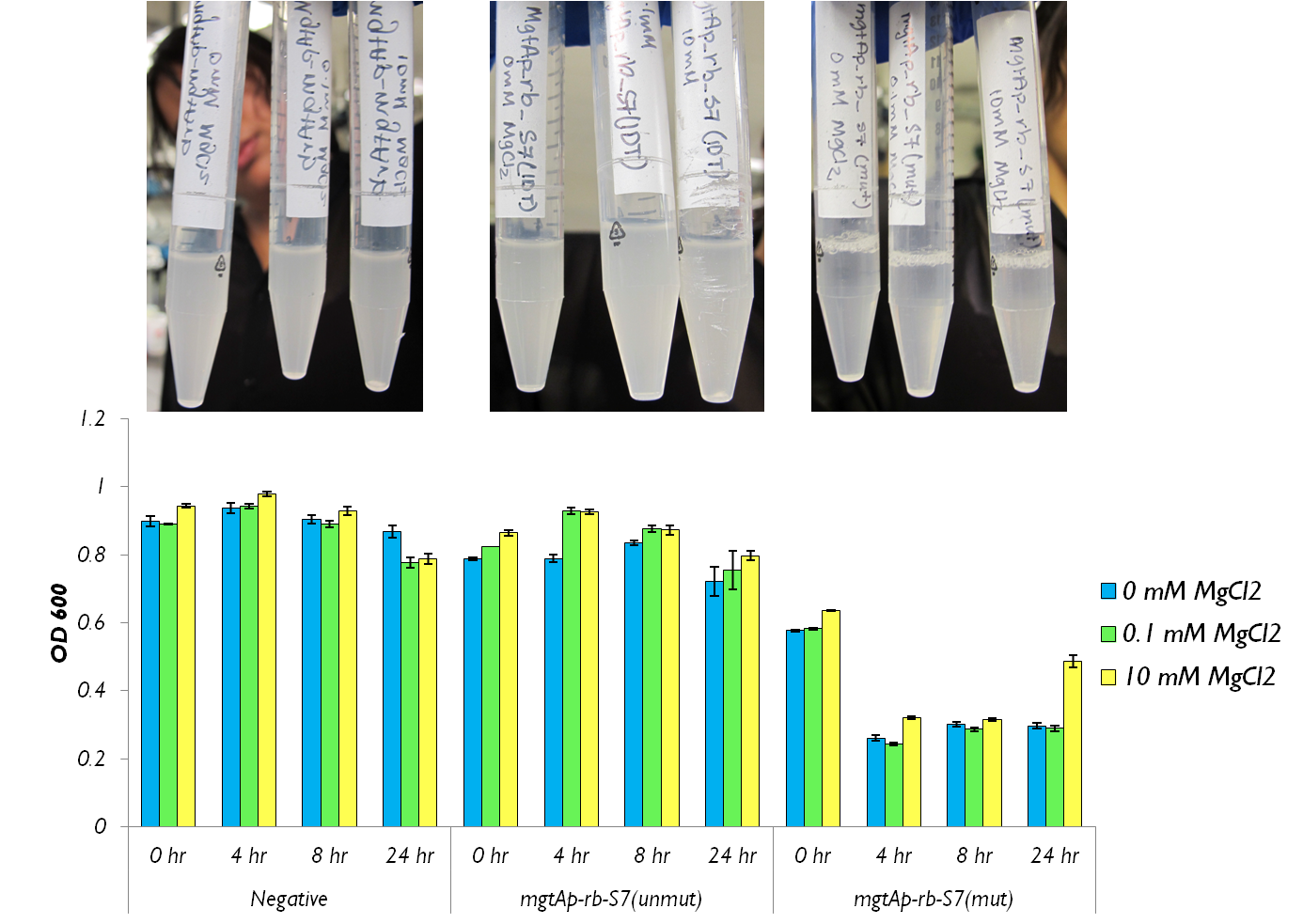
In tandem to building our systems with GFP in order to test their control, we also constucted them with our kill genes. This was delayed substantially however due to problems in their synthesis. Specifically, the micrococcal nuclease that arrived from IDT had a 1bp mutation which changed a isoleucine residue into a lysine. Initially, our systmes resulted in no killing of cells. Therefore we had to mutate this resiude using site-directed mutagenesis. Once completed, we were able to begin testing. With our GFP data collected, we moved on to characterizing the mgtA control system upstream of our S7 kill gene (BBa_K902019). To test the circuits, we incubated cells expressing our construct with differing concentrations of magnesium. We then measured both Colony forming units and OD 600. For a deatiled protocol, see here.
Results
Figure 6 shows that the mgtAp-mgtArb-S7 (BBa_K902018) starts acting approximately 4 hours after induction. However, it also shows that 10mM MgCl2 is not enough salt to inhibit the entire system because there is no difference in OD600 measurement at 4hr timepoint between 10mM and the 0mM concentrations. This test needs to be repeated with higher concentrations of Mg2+ however this data suggests that the mutagenesis was successful and S7 is active and killing the cells at approximately 4hr which does not necessarily reflect upon the activity of S7 but also reflects upon the response time of the mgtA system.
An alternative: a glucose repressible system
Based on the problem with the magnesium and system in relation to tailings ponds, we wanted to find an alterntative (other than the manganese and moco systems, which require further testing) We found a promoter in the literature that was both induced by rhamnose and repressed by glucose. This seemed to be a very suitable candidate for controlling the kill switch in the the bioreactor since the promoter is shown to be tightly repressed by glucose and rhamnose are fairly inexpensive and not found in high concentrations in tailings ponds. We could supplement the bioreactor with glucose to inhibit expression of the kill genes in the bioreactor. Escape of bacteria into the tailings ponds will cause expression of the kill genes since glucose levels in the surrounding environment would be insufficient for deactivate the promoter.
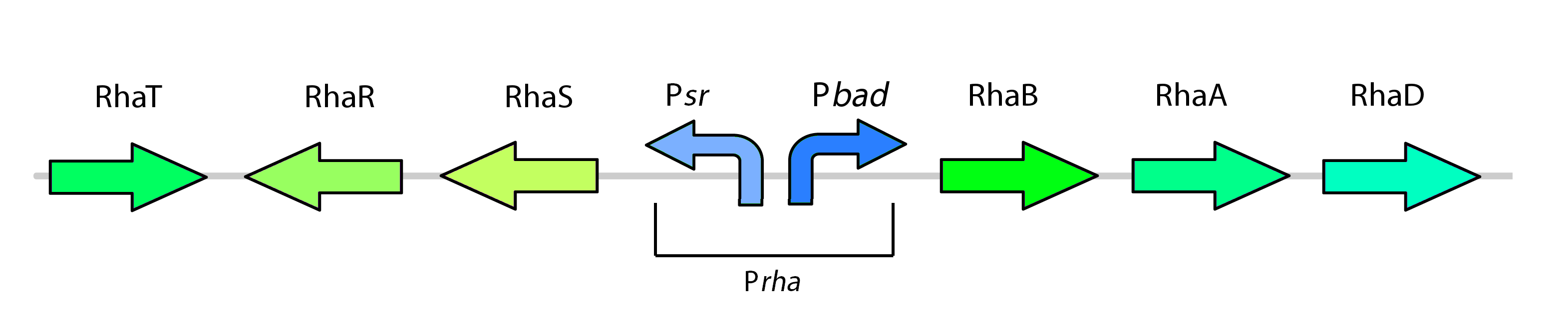
This promoter, known as pRha (BBa_K902065), is responsible for regulating six genes related to rhamnose metabolism and contains a separate promoter on its leading and reverse strands (see figure 7). RhaR (BBa_K902069) and RhaS (BBa_K902068) serve to regulate expression of the rhamnose metabolism genes rhaB, rhaA, and rhaD on the opposite side of the promoter. The RhaR transcription factor is activated by L-rhamnose to up-regulate expression rhaSR operon. In turn, the resulting RhaS activates the rhaBAD operon to generate the rhamnose metabolism genes (Egan & Schleif, 1993).
As a kill switch regulator, our team has harnessed global catabolite repression of the rhamnose promoter. Expression of the rhaBAD operon with RhaS requires the binding of catabolite receptor protein (CRP) cAMP complex to the promoter. When glucose is present, cAMP levels are low, such that CRP is unable to activate the promoter (Egan & Schleif, 1993). With this mechanism, our team set out to control our kill genes with glucose with the following rhamnose promoter construct:
Our kill system is different from the native rhamnose system with the rhaR and rhaS control genes. We have constituitively expressed RhaS to overcome dependency on rhamnose to cause activation of the kill switch. While RhaS is continuously present, global catabolite repression prevents its activation the kill genes in the bioreactor; in the outside environment, glucose levels are lower such that RhaS is able to activate the kill genes.
Building the system
Our team had pRha promoter (BBa_K902065) commercially synthesized as per the sequence given by Jeske and Altenbuchner (2010). The rhaS (BBa_K902068) and rhaR (BBa_K902069) genes were amplified via PCR from Top Ten E. coli using Kapa Hi-Fi polymerase.
Given the control gene modifications which we have engineered into our system to optimize it for the tailings ponds, we are working to determine whether glucose repression of our modified system can match patterns shown by Giacalone et al. (2006) and Jeske and Altenbuchner (2010). To this end, we have constructed the rhamnose promoter with GFP (BBa_K902066) and are finalizing construction with constituitively expressed RhaS, so that we can characterize this promoter and test it in combination with our kill genes.
 "
"