Team:LMU-Munich/Germination Stop

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


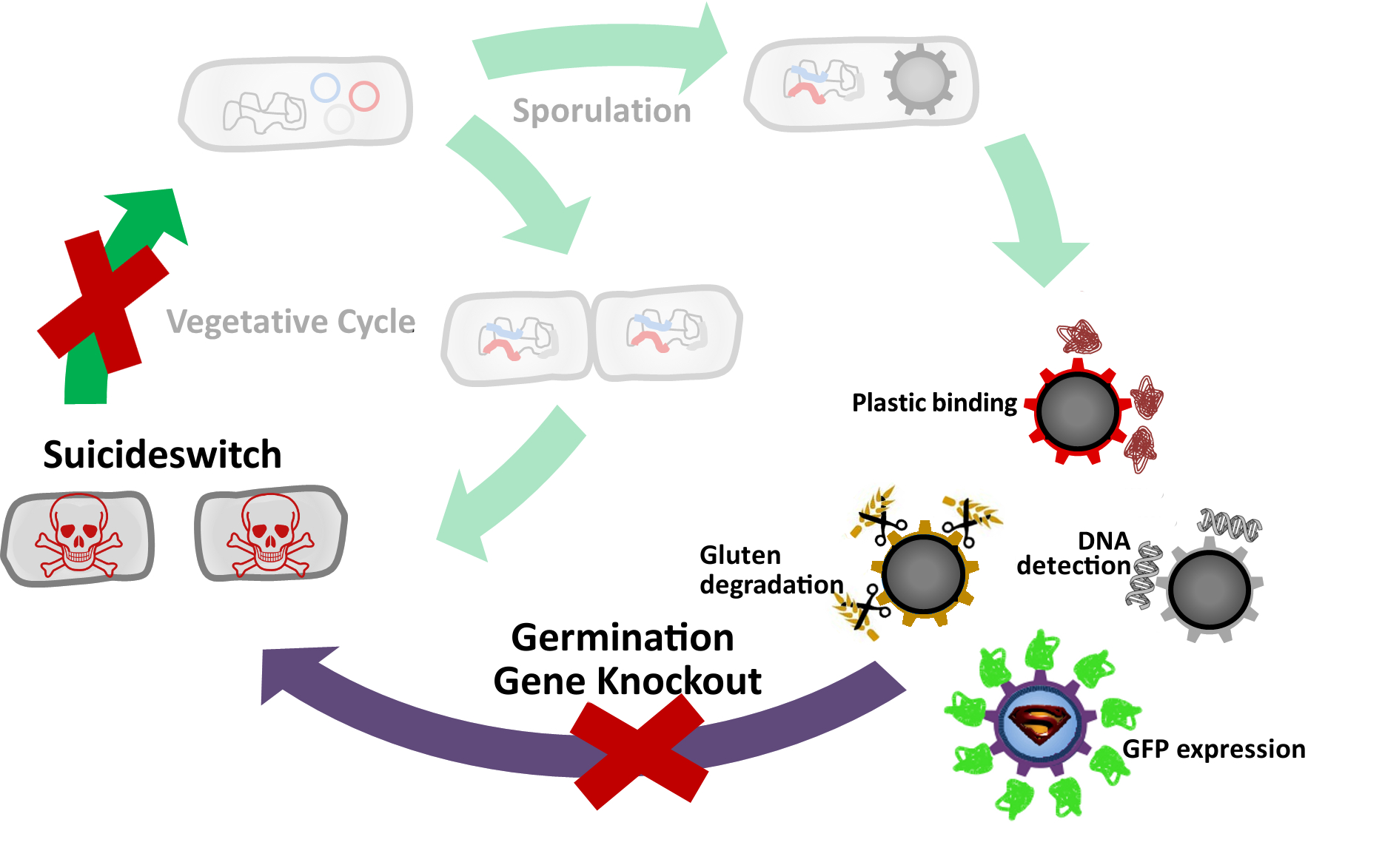
GerminationSTOP
The goal of this module was to yield Sporobeads which are safe (unable to germinate) and consistently functional (maintain their spore shape and structure throughout time). To achieve this, we sought to remove the germination capability of our spores, while keeping their necessary structural functions intact.
Two approaches were used to achieve this:
- Knock out genes that are involved in germination.
- Suicideswitch: Toxin production by vegetative cells if germination knockout fails and spores manage to germinate.
How do Sporulation & Germination Work?
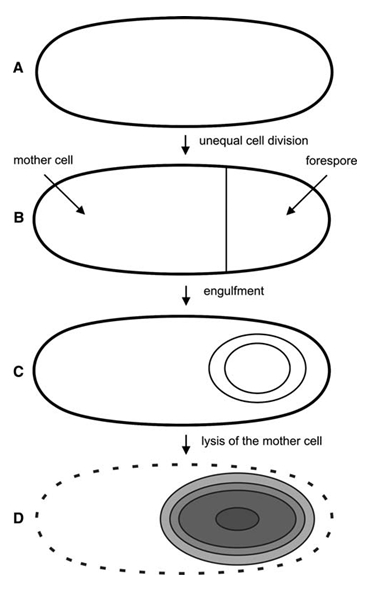
Sporulation: The Bacillus life cycle can include both classic division as well as reproduction by sporulation and spore germination. In response to nutrient starvation, Bacillus cells form endospores in a process called sporulation (Fig. 1). The “mother” cell forms the endospore within its own cytoplasm. The endospore contains its DNA in the spore core, which is protected by several layers of coats. The outermost layer is the spore crust. The spore is very dry, and contains a substance called dipicolinic acid (DPA), which replaced water to protect the DNA. Until the spore hydrates and swells out of its protective coats, it is resistant to a wide variety of environmental stressors, including UV radiation, toxic chemicals, freezing, high heat, dessication, and pH extremes. This resistance to stressors allows the spore to survive almost indefinitely until conditions are again ideal for growth.
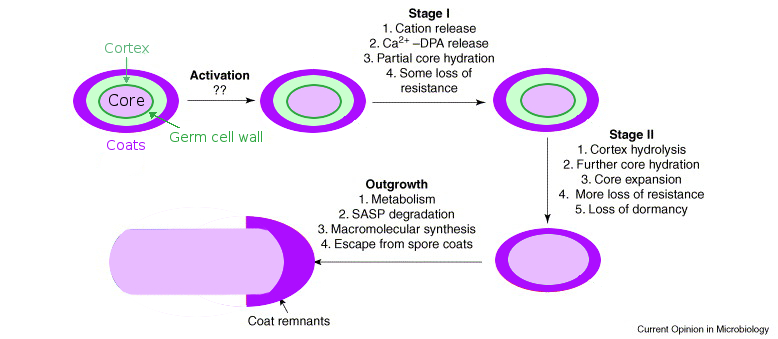
Germination: On its inner spore membrane, the spore has germinant receptors. The spore coats are believed to be semipermeable or porous, in order to permit the passage of germinants to the receptors. When germinants such as amino acids and sugars reach the receptors, the spore begins the biophysical process of germination (see Fig. 2). Initially, the spore replaces its DPA with water (Stages I and II in Fig. 2), shifts its pH (Stage I), and swells (Stage II). Lytic enzymes break down the rigid spore coat layers encapsulating the spore's core (Stage II), allowing the spore core to swell further and begin the outgrowth process into a vegetative cell (Outgrowth). When the spore coat is broken down by lytic enzymes, the spore's environmental resistance is lost. For our project, we wish to prevent this germination process.
|
How do Germination Gene Knockouts Work?
Our idea of how to prevent germination from occurring is to knock out genes encoding functions involved in the germination process. There are a huge number of genes which play a role in germination, but our goal was to select a few very essential genes to knock out.
Based on the research of others, the genes we chose were cwlJ, sleB, cwlB, gerD, and cwlD. Past works of [http://www.ncbi.nlm.nih.gov/pubmed/19554258 J. Kim and W. Schumann (2009)] and [http://www.ncbi.nlm.nih.gov/pubmed/11466293 B. Setlow et al. (2001)] showed:
- cwlJ and sleB: when knocked out together, germination frequency was reduced by 5 orders of magnitude. Both genes code for lytic enzymes which are active in the process of germination.
- gerD and cwlB: When knocked out together, germination frequency was reduced by 5 orders of magnitude. The gerD product plays an unknown role in nutrient germination; the product of cwlB plays a role in cell wall turnover & cell lysis. Note: after several attempts to knock out cwlB, and yielding cells unable to grow or cells growing at a very slow rate, we decided that knocking out cwlB would not be prudent for the production of Sporobeads, as it would delay all experiments in the GerminationSTOP module.
- cwlD: When knocked out, germination occurred at a rate of 0.003 to 0.05%. This gene codes for recognition components for cleavage by the germination-specific cortex lytic enzymes.
By knocking out all five of these genes, our goal was to yield a B. subtilis strain which produces spores completely incapable of germination. Germination is stopped during the process of spore coat breakdown. Without the lytic enzymes to break down the coat, the spores should be unable to outgrow into the vegetative stage.
Table 1: Literature values extracted from [http://www.ncbi.nlm.nih.gov/pubmed/19554258 J. Kim and W. Schumann (2009)] and [http://www.ncbi.nlm.nih.gov/pubmed/11466293 B. Setlow et al. (2001)].
| Germination Genes | Encoded Function | Mutant Germination Rate |
|---|---|---|
| gerD | Unknown role in nutrition germination | Reduction by 5 orders of magnitude |
| cwlJ | Germination-specific lytic enzymes | 0.003 – 0.05%
No ATP detected |
| sleB | Germination-specific lytic enzymes | 0.003 – 0.05%
No ATP detected |
| cwlD | Recognition component for lytic enzymes | 0.003 – 0-005% |
Germination Gene Knockout Success!
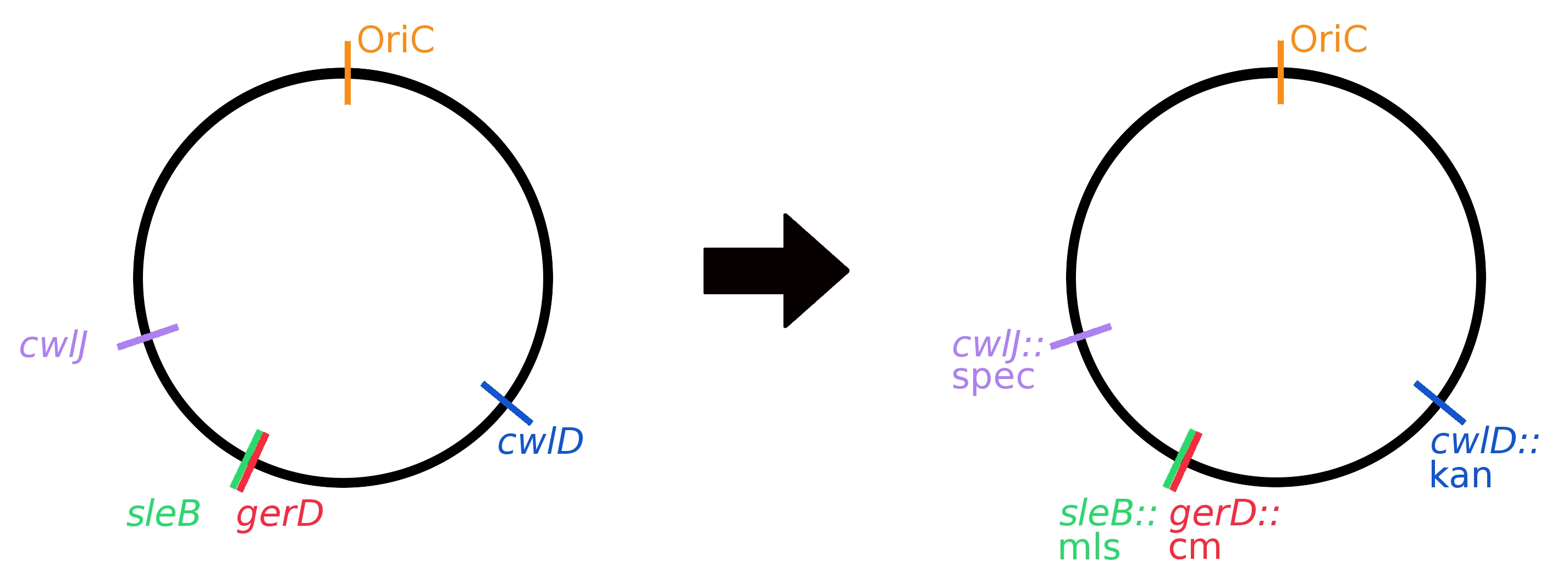
Two methods were employed to knock out germination genes: replacement by resistance cassettes and clean deletions. Resistance cassette knockouts were generated by using long-flanking homology PCR (see Protocols). Single knockouts were created first using different resistance cassettes; then they were combined to create multiple knockouts. A graphical representation of the genome with all gene replacements can be seen in Fig 3.
|
The germination rate of our mutants was checked with a germination assay, shown in Fig. 4. The assay was developed based on published protocols ([http://www.amazon.co.uk/Molecular-Biological-Methods-Bacillus-Microbiological/dp/0471923931/ref=sr_1_1?ie=UTF8&qid=1348709578&sr=8-1 Harwood, Cutting (1990) Molecular Methods for Bacillus subtilis. Wiley].
|
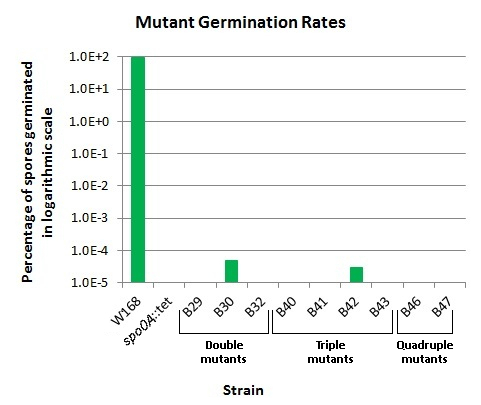
We were able to achieve germination rates as low as <1 in 4.6x109 spores in our triple mutant B43. Other mutants also showed extremely reduced germination ability. A table of germination rates of our mutants and all of the other spectacular results can be found on our results page. Our single mutants were not checked in the germination assay. The double and triple mutants tested showed mixed results, with some unable to germinate, and others maintaining low germination abilities. Our quadruple mutants demonstrated no ability to germinate, even in several separate germination assays.
Despite this success, safety was a top priority for this project. In the event that some small percentage of spores retained the ability to germinate, the Suicideswitch subproject (below) prevents outgrowth into viable vegetative cells.
Suicideswitch
|
As a backup plan to make our Sporobeads even safer, we developed the Suicideswitch. In case the spores do germinate, due to degradation or destruction of their outer coats, e.g. by high pressure, the Suicideswitch will be turned on.
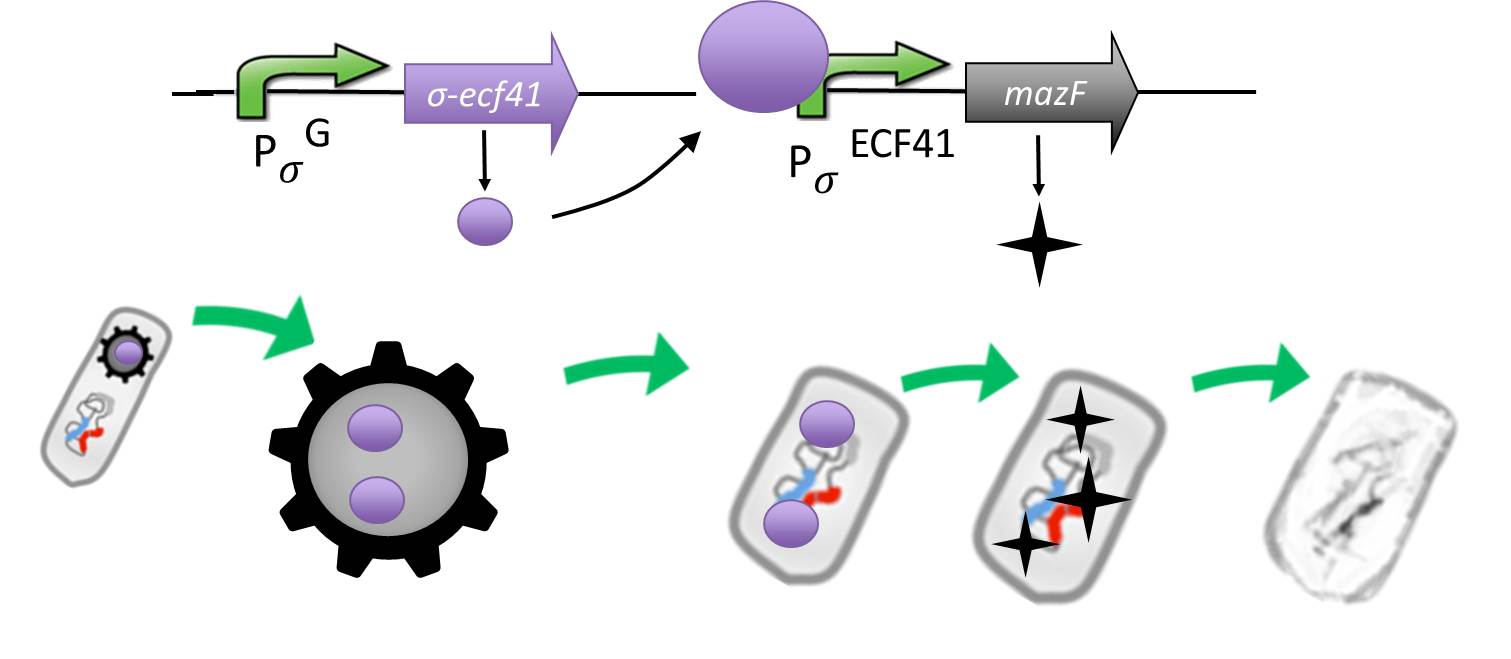
We take advantage of an alternative and heterologous σ factor and the target promoter. Both are not present in B. subtilis and therefore not recognized by any other σ factor of B. subtilis.
The alternative sigma factor [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823043 ecf41Bli aa1-204] ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3426412/ Wecke et al. 2012]), which derives from B. licheniformis a truncated version constitutively "ON" is synthetically linked to one out of two σG-regulated promoters ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823048 PspoIVB] ([http://www.ncbi.nlm.nih.gov/pubmed/15699190 Steil et al. 2005]) responding strongly or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823042 PsspK] ([http://www.ncbi.nlm.nih.gov/pubmed/15699190 Steil, Völker et al. 2005]) responding weakly). σG is the last σ factor activated in the forespore. Consequently, Ecf41Bli aa1-204 is produced quite late and only in the forespore. Ecf41Bli aa1-204 then activates its unique target promoter [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823041 PydfG] ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3426412/ Wecke, Mascher 2012]). This promoter is fused to [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823044 MazF] ([http://jcs.biologists.org/content/118/19/4327.abstract Engelberg-Kulka, Amitai 2005]). Activation of PydfG therefore leads to the expression of MazF, a bacterial toxin from E.coli degrading mRNA.
The resulting time course of expression is as follows: Sporulation of the Sporobeads leads to Ecf41Bli aa1-204 production in the forespore at the end of the differentiation cycle, through the activation of either PspoIVB or PsspK. Subsequently, this would lead to the expression of MazF, due to the activation of PydfG. For our already matured Sporobeads, this would not be toxic, as they represent a dormant state that does not require translation any more. But in the case that a Sporobead escapes the GerminationSTOP based on the gene knockouts described above, a germinating cell will already contain or soon start producing MazF and therefore kill the vegetative cells.
See what Data we got when measuring a module with luxABCDE instead of MazF.
We are planning to model this system, but still need more quantitative, time-resolved data for it. We hope to get this data after finishing the cloning of PsspK and MazF into B. subtilis.

| 
| 
| 
|
| Bacillus Intro | Bacillus BioBrickBox | Sporobeads | Germination STOP |
 "
"