Team:SEU O China/HumanPractice
From 2012.igem.org


Human Practice
Safety
We appointed biosafety as our theme for human practice. As a very significant part of our project, several activities have been conducted to fulfill our arrangements.
First of all, we carefully examined the details of the biosafety questionnaire and decided to conduct a survey on the biosafety issue at our own school. Observations were made during our regular experiment process over this issue and we’ve found out several fundamental requirements as well as limitations on our laboratories. With the supervision of several professors and graduates, we got some basic ideas on lab biosafety issue even at the very beginning of our experiment process. One key point lies in the separation of bacterial, DNA and RNA experiments, which should be essential to modern integrated biological laboratories. Adding to that is so basic a point that it is often neglected by us, and that is a proper classification of preserved bacteria as well as a compact remark on the lab items. The neglect would easily evolve into pollution and destroy the experiment. The limits, however, are obvious, due to limited funds and conditions, some disposable equipment are in fact infected and reused several times. Also, the aseptic technique is not always met up with the standard. In conclusion, though with the reference to a relatively standard biosafety standard, there is still some place to improve in our lab process.
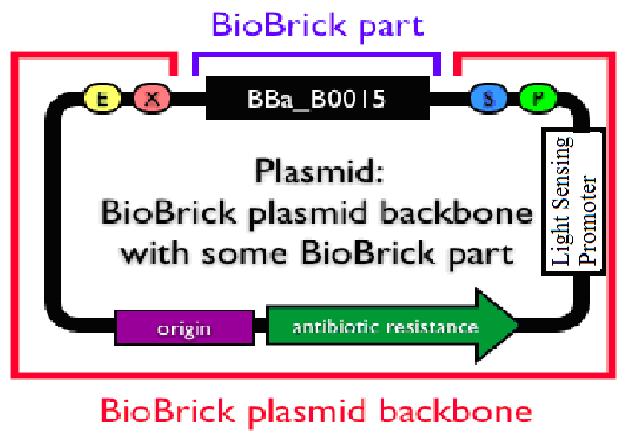
Secondly, in improving the actual safety standard in modern biology, we try to construct a new general transgenic vector. The principle is rather simple: using light condition to manipulate the growth of colony. We have successfully constructed a rather high efficient division repression biobrick and we design a special part consisting of a light sensor and this division repression biobrick. Unfortunately, the fabrication of this small system has not been finished yet due to lack of time. But once the connection is successful, a promising general biological tool would emerge. Imagine the circumstance where the colony is exposed to certain length of light and then died automatically. What the experiment executers need to do is to add specific light (physical) condition to the colony. When the experiment finished, an exposure to normal solar light would directly make the colony die and prevent pollution from the laboratory and therefore improve the biosafety standard.
In the final analysis, our human practice is focused on biosafety issue but also put some attention on the improvement of this general subject as well as this competition. We are just trying to combine what we are doing as our system with the improvement of biosafety standard.
Safer Transgenic Vector
In improving the actual safety standard in modern biology, we try to construct a new general transgenic vector. The principle is rather simple: using light condition to manipulate the growth of colony.
What we are expecting to get is a newly-built plasmid backbone. In addition to an antibiotic resistance gene for selecting, add in a simple light sensor expressing path would be added in.
Two schemes might be possible for this design.
Scheme 1:
Just add a light sensor promoter on the upstream of restriction site. This is a rather simple scheme which allows the plug-in genes to express under a certain light condition. However, to express plug-in genes, a RBS must be ligated in at the same time. Moreover, this kind of procedure can just be equal to using light to start a process. Although such a process may seem to be controlled by light, it should be noted that normal solar light contain almost all lengths of visible light and some common invisible light. Even white lamp would limit the meaning of this part. So this design is not rather ideal.
Scheme 2:
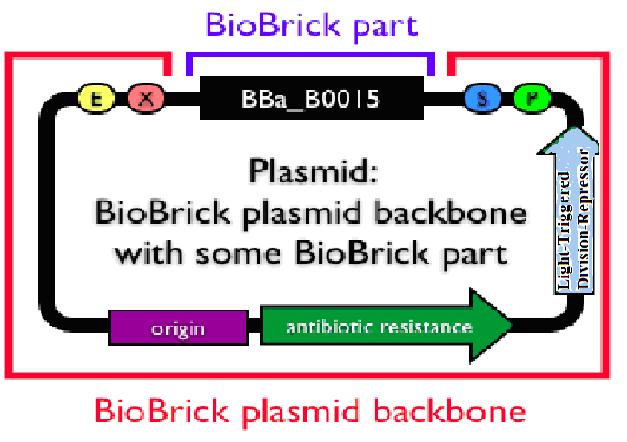
Now that the first design is not practically possible, consider the problem from the back side.That is, make the plug-in genes express only when there is no light.The principle is quite simple. Attach another division repressing part to the former light sensor and make the transformed bacteria stop growing when there is a certain length of light. We have successfully constructed a rather high efficient division repression biobrick and we design a special part consisting of a light sensor and this division repression biobrick. As the figure on the right indicates, a light-triggered division-repressor could be constructed and added in the backbone. One option might be the division repression biobrick we designed ourself.
Unfortunately, the fabrication of this small system has not been finished yet due to lack of time. But once the connection is successful, a promising general biological tool would come out. Imagine the circumstance where the colony is exposed to certain length of light and then died automatically. What the experiment executers need to do during the transgenic process is to add specific light (physical) condition to the colony. When the experiment finished, an exposure to normal solar light would directly make the colony die and prevent pollution from the laboratory and therefore improve the biosafety standard.
Video
In August, we issued a short flash video named Synthetic Biology---yesterday, today and tomorrow to promote the knowledge of synthetic biology. This about 3 minute-long video briefly introduces the history, basic principle, standard procedures and potential applications of this modern subject.
The video can be seen [http://www.youtube.com/watch?v=HRsescThcmA&feature=youtu.be here].
Relevant Information:
The term "synthetic biology" has a history spanning the twentieth century.[1] The first use was in Stéphane Leducs’s publication of « Théorie physico-chimique de la vie et générations spontanées » (1910) [2] and « La Biologie Synthétique » (1912).[3] In 1974, the Polish geneticist Wacław Szybalski used the term "synthetic biology",[4] writing:
Let me now comment on the question "what next". Up to now we are working on the descriptive phase of molecular biology. ... But the real challenge will start when we enter the synthetic biology phase of research in our field. We will then devise new control elements and add these new modules to the existing genomes or build up wholly new genomes. This would be a field with the unlimited expansion potential and hardly any limitations to building "new better control circuits" and ..... finally other "synthetic" organisms, like a "new better mouse". ... I am not concerned that we will run out of exciting and novel ideas, ... in the synthetic biology, in general.
When in 1978 the Nobel Prize in Physiology or Medicine was awarded to Arber, Nathans and Smith for the discovery of restriction enzymes, Wacław Szybalski wrote in an editorial comment in the journal Gene:
“The work on restriction nucleases not only permits us easily to construct recombinant DNA molecules and to analyze individual genes, but also has led us into the new era of synthetic biology where not only existing genes are described and analyzed but also new gene arrangements can be constructed and evaluated.”
By now, synthetic biology has developed a standard execution procedure:
BioBrick
Standard biological parts are DNA sequences of defined structure and function; they share a common interface and are designed to be composed and incorporated into living cells such as E. coli to construct new biological systems. BioBrick parts represent an effort to introduce the engineering principles of abstraction and standardization into synthetic biology. The trademarked words BioBrick and BioBricks are correctly used as adjectives (not nouns) and refer to a specific "brand" of open source genetic parts as defined via an open technical standards setting process that is led by the BioBricks Foundation.
Restriction Digest&&DNA Ligation The restriction digest technique can be used for cleaving DNA molecules at specific sites, ensuring that all DNA fragments that contain a particular sequence have the same size; furthermore, each fragment that contains the desired sequence has the sequence located at exactly the same position within the fragment. The cleavage method makes use of an important class of DNA-cleaving enzymes isolated primarily from bacteria. These enzymes are called restriction endonucleases or restriction enzymes, and they are able to cleave DNA molecules at the positions at which particular short sequences of bases are present. DNA ligation is the process of joining together two DNA molecule ends (either from the same or different molecules). Specifically, it involves creating aphosphodiester bond bond between the 3' hydroxyl of one nucleotide and the 5' phosphate of another. This reaction is usually catalyzed by a DNA ligase enzyme. This enzyme will ligate DNA fragments having blunt or overhanging, complementary, 'sticky' ends. Typically, it is easier to ligate molecules with complementary sticky ends than blunt ends. T4 DNA ligase is the most commonly used DNA ligase for molecular biology techniques and can ligate 'sticky' or blunt ends.
DNA sequencing
DNA sequencing is determining the order of the nucleotide bases in a molecule of DNA. Synthetic biologists make use of DNA sequencing in their work in several ways. First, large-scale genome sequencing efforts continue to provide a wealth of information on naturally occurring organisms. This information provides a rich substrate from which synthetic biologists can construct parts and devices. Second, synthetic biologists use sequencing to verify that they fabricated their engineered system as intended. Third, fast, cheap and reliable sequencing can also facilitate rapid detection and identification of synthetic systems and organisms.
PCR
The polymerase chain reaction (PCR) is a biochemical technology in molecular biology to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence.
Transgenesis
Transgenesis is the process of introducing an exogenous gene – called a transgene – into a living organism so that the organism will exhibit a new property and transmit that property to its offspring. Transgenesis can be facilitated by liposomes, plasmid vectors, viral vectors, pronuclear injection, protoplast fusion, and ballistic DNA injection.
Measurement
Precise and accurate quantitative measurements of biological systems are crucial to improving understanding of biology. Such measurements often help to elucidate how biological systems work and provide the basis for model construction and validation. Differences between predicted and measured system behavior can identify gaps in understanding and explain why synthetic systems don't always behave as intended. Technologies which allow many parallel and time-dependent measurements will be especially useful in synthetic biology.
Prospect:
Bacteria Photo; Bio-Toggle Switch; Synthetic Life; Bio-factory; Bio-computing; Bio-fuel;
Reference:
[1]James Collins,2012. Bits and pieces come to life. Nature, volume 483; [2]http://en.wikipedia.org/wiki/Synthetic_biology; [3]http://en.wikipedia.org/wiki/Restriction_digest; [4]http://openwetware.org/wiki/DNA_ligation.
Entertainment
In the rest the experiment time, we surely have a lot of entertainment. Having two senior MTG fan in our group, card game becomes a important part of our entertainment time. Beside playing cards, we designed several new cards ourselves, most of which reflect the problems and obstacles we meet during experiments. Here are some of them:
Others
It is the first year for Southeast University to join iGEM competition. Since several member of us heard of iGEM last autumn, we put great efforts in persuading the professors for support and raising fund from school, university and government project. It is already a huge success for us that we finally organize the school team and march towards the regional jamboree.
We organized a lecture on February to introduce the iGEM to students, and held a school competition to recruit members for the team on March. The school competition last a week and consist four parts: scheme design, mathematical modeling, biological experiment and website design. We also invite professors in related subject of our school as judges. The students who attend the competition and have interest in the project was recruited in our team.
The result of our school competition can be seen [http://jwc.seu.edu.cn/s/99/t/1600/e7/47/info59207.htm here] (Chinese version only).
- The presentation of scheme design part
In September, we held a lecture in the university with the other team of our school, SEU_A, and the two teams of Nanjing University, to present our projects and show the idea of synthesis biology.
- The synthesis biology lecture

 "
"