Team:Marburg SYNMIKRO/Results
From 2012.igem.org

Results
Biobricks:
BBa_K866000 [Link] (CFP-Fusion Brick):
In order to evaluate putative recombination events in E. coli, we chose to generate fluorescent fusion proteins which are to be localized at different locations within the cell. BBa_E0020 [Link] is a biobrick coding for the Cyan Fluorescent Protein (CFP). To build a CFP suitable for fusion proteins, we chose this biobrick as a template to create a CFP coding sequence (CDS) which can be cloned in-frame to other protein coding sequences via Standard Assembly [Link]. The CFP-Fusion Brick was constructed through site directed mutagenesis by polymerase chain reaction (PCR) using primer MS_16 [Link] and MS_17 [Link]. PCR products were separated by agarose gelelectrophoresis, gel extracted and cloned into pSB1C3 [link] via Standard Assembly [link]. Positive clones have been verified by colony PCR [link colony-pcr-bild in lab-book?] and sequencing [link sequenzierung in parts registry?]. This fusion brick was later on screened for expression and localization in E. coli as a synthesized fusion protein with A-modules GroES [link and AmiC [link] using fluorescence microscopy [link zu fluoreszenz-experiment].
BBa_K866001 [Link] (mRFP-Fusion Brick):
Besides a CFP-fusion protein, a fusion “competent?” monomeric Red Fluorescent Protein was contructed from BBa_E1010 [Link] as template. Site directed mutagenesis was performed via PCR using primer MS14[Link] and MS15 [Link]. PCR products were separated by agarose gelelectrophoresis, gel extracted and cloned into pSB1C3 [link] via Standard Assembly [link]. Positive clones have been verified by colony PCR [link colony-pcr-bild in lab-book?] and sequencing [link sequenzierung in parts registry?].
BBa_K866002 [Link] (G-segment Invertase): The G-segment Invertase (Gin) from the phage Mu [Link?] is a protein which can either cause inversions of gene segments flanked by gix-sites [link paper oder Gene bank, da keine sequenz in parts registry?] as inverted repeats or cause deletions of gene segments flanked by gix-sites as direct repeats [link paper bzw. ,,recombination”-grundlagen in unserem wiki?]. Recombination events are facilitated by the Enhancer DANN segments located in cis [link paper?] (see also poste/project [link]). We were aiming at constructing a biobrick that codes for Gin [link] and contains a ribosomal binding site (RBS) attached/flanked upstream from it’s CDS.
Hence, we designed primer (MS11 and MS12 [link]) which result in amplification of Gin containing the well characterized and strong RBS BBa_J61100 [link registry] from the Anderson RBS-family. As template for PCR, chromosomal DNA from E. coli strain C600 Mucts62 (lysogenic for temperature sensitive phage Mu cts62) was used. PCR amplicons were separated via agarose gelelectrophoresis and cloned into pSB1C3 through Standard Assembly [link]. Positive clones were evaluated by colony-PCR [link labbook] and sequencing [link parts registry???]. The G-Segment Invertase Biobrick [link] can be tested with different promoters in order to evaluate the optimal level of expression for recombination events in E. coli to occur.
BBa_K866002 [Link] (SacB Suicide Gene): In order to select for recombination events in E. coli cells in a culture, the suicide gene SacB [link] was chosen. In our final recombination construct [link project], it shall be located between A and B-modules, causing a deletion of this suicide gene and thus select for positive recombination events. It codes for a levan sucrase, which can polymerize sucrose from growth media within Gram negative cells to levan, thus disturbing the cell’s metabolism and hence growth, leading to growth inhibition up to cell death [link paper?]. SacB was amplified using primer MS26 and MS27 [link primer liste] from the suicide plasmid pNTPSXY via PCR. PCR products were separated by agarose gelelectrophoresis, gel extracted and cloned into pSB1C3 [link] via Standard Assembly [link]. Positive clones have been verified by colony PCR [link colony-pcr-bild in lab-book?] and sequencing [link sequenzierung in parts registry?]. BBa_K866002 [link] can be fused to promoters of different strength and tested for optimal screening through cell death on sucrose rich media.
Experiments:
Generation of test constructs: As a first step towards our complete recombination construct [link poster, bzw. Project], we aimed at generating the putative recombination gene products, meaning the fusion of a complete A-module with a complete B-module with a gix-site in between. These putative recombination constructs would be the outcome as if a recombination event would have had occurred in the complete construct [zu viel “construct” -> alternatives wort???]. For this purpose, all three complete (HU nicht dabei, trotzdem alle 3 erwähnen?) A-Modules, HU [link], AmiC[link] and GroES[link] have been synthesized by Gene Art (life technologies TM). These A Modules consist each of one localization domain (AmiC, GroES or HU) with the RBS BBa_J61100 [link] and the strong constitutive promotor BBa_J23108 [link] located upstream. Downstream of localization domains, a gix site [link] is located. Between promotor, RBS, localization domain and the gix site, the scar sequence from the standard assembly [link scar] was included in order to test for the correct reading frame as it would have been necessary for the complete recombination construct. The complete A-modules are flanked by the standard prefix and suffix. The complete A-modules coding for AmiC and GroES were cloned N-terminally to our fluorescent fusion protein sequences BBa_K866000 (CFP-Fusion Brick) [link], BBa_K866001 (mRFP-Fusion brick) [link] aswell as the biobrick BBa_K125500 (GFP-Fusion Brick) [link] via standard assembly [Link]. Verification of fluorence was performed via fluorescence microscopy.
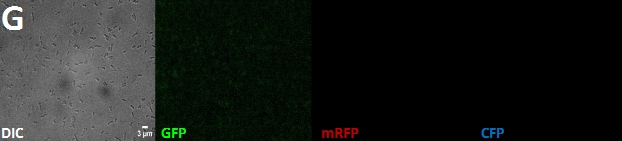
Fluorescence microscopy: In order screen for E. coli containing and expressing our test constructs, positively selected colonies through antibiotic restistance have been subjected to fluorescence microscopy. For this purpose, colonies growing on LB-Agar supplemented with antibiotics for BBa_K866000 (CFP-Fusion Brick) [link], BBa_K866001 (mRFP-Fusion brick) [link] and BBa_K125500 (GFP-Fusion Brick) [link], respectively, were cultured over-night in LB-medium with antibiotics. Small aliquots (10 µL) of overnight cultures were added onto 2% agarose-pads [link protokoll, falls überhaupt vorhanden?] located on microscope slides, covered by cover-slips (doppelt gemoppelt oO) and used for fluorescence microscopy.
Alltogether, 6 different constructs have been observed for fluorescence, being AmiC-GFP, AmiC-CFP, AmiC-mRFP, GroES-GFP, GroES-CFP and GroES-mRFP. As visible in figure XY, fluorescence occurs in cell carrying each construct. Especially in cells expressing GroES-GFP and AmiC-GFP figure XY, intense fluorescence is detectable at cell poles and in the periplasm in contrast to other cell compartments. Similar effects are also visible for GroES-mRFP and AmiC-mRFP, respectively. For GroES-CFP and AmiC-CFP figure XY, fluorescence is detectable, yet no precise localization can be determined.
As a result, our test constructs mimicking (???) a putative recombination event of our planned complete recombination construct [link projet] are expressed in E.coli. Furthermore, GFP- and mRFP fused A-modules show enhanced fluorescence at expected cell compartments, being cell poles for GroES and periplasm for AmiC.
Discussion:
Outlook:
 "
"