Why?
From our work on LOV2 photoactivation we might be able to predict the percentage of LOV2 domains in our LovTAP-VP16, even though this value might be affected by the fact of having both ends fused to other domains: TrpR and VP16. However, some authors, as Kasahara et al, worked with fusion LOV2 proteins, getting comparable parameters.
How?
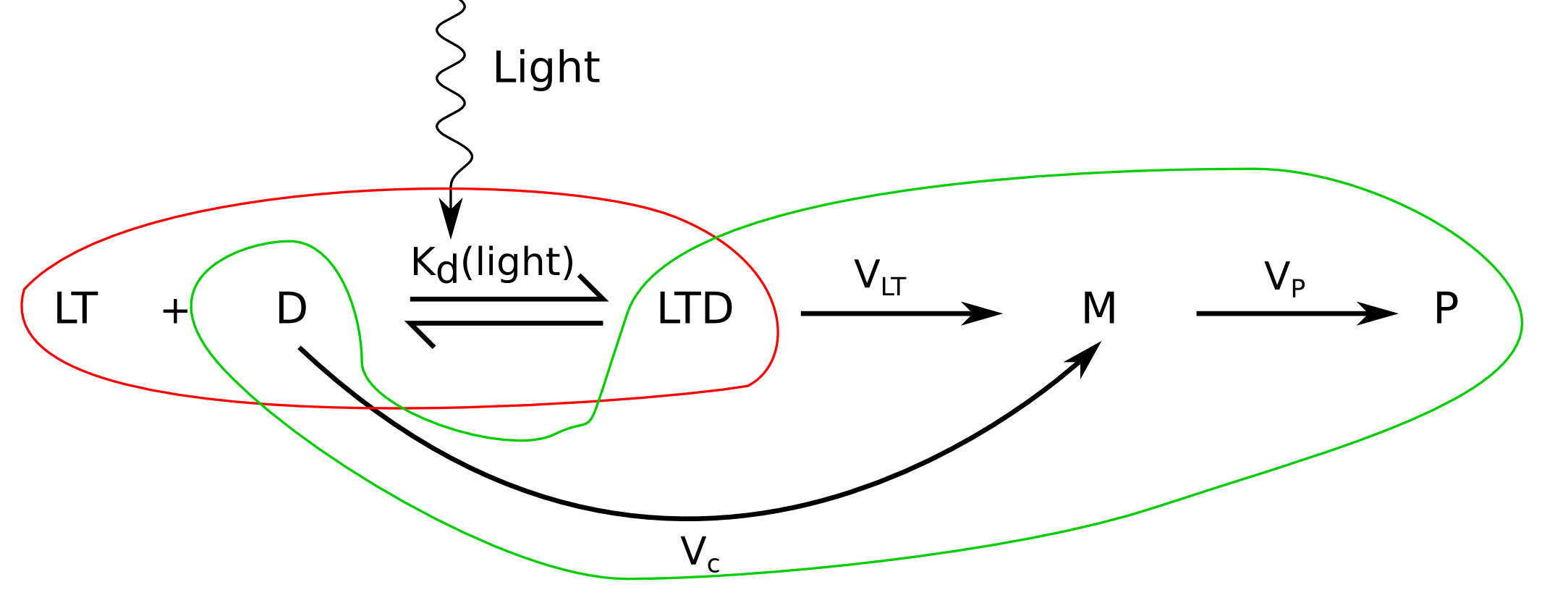
Photoactivation level
We will then assume that we know the proportion of activated LovTAP-VP16 monomers, since we have calculated it in this section. But LovTAP-VP16 has to dimerize to be able to bind DNA. To illustrate the importance of this, let's give an example: imagine a solution with LovTAP-VP16 where 50% of the monomers are activated. Assuming that all of them form part of a dimer, we get that 25% of the dimers are inactive, 25% active and 50% half active. The average binding affinity will mostly depend on the affinity of the half active half inactive dimers. In the section on the 3D protein structure of LovTAP-VP16 we conclude that it is reasonable to think that a half activated dimer will have a DNA activity somewhere between a fully activated dimer and a deactivated one. This means that we can take the monomer activation level to approximate the dimer activation level.
92% of photoactive LOV2 has it's Jα-helix undocked (Yao et al, 2008).
Eitoku et al (2005) say it takes 2 ms for the J&alpha-helix to undock.
 "
"