Team:Marburg SYNMIKRO/Project
From 2012.igem.org

Summary
We aimed to establish a system in Escherichia coli cells that generates a large number of novel proteins by combinatorial fusion of subfragments. Our project was inspired by the VDJ-recombination of the vertebrate immune system. There, a limited number of protein coding sequences is used to generate the enormous diversity of antibodies. We designed A and B modules containing different protein coding subfragments to be recombined. Fusion of one A fragment with one B fragment is achieved by the site-specific DNA recombinase Gin of bacteriophage Mu. Recombination sites were arranged as direct repeats thus causing deletions upon recombination. Gin catalyzed recombination depends on the presence of an enhancer element.This enhancer was placed together with the suicide gene sacB between the A and B modules. This design guarantees that recombination automatically stops when the central fragment is deleted. To test our system and to visualize the combinatorial activity we fused fluorescent proteins of different color to different intracellular localization domains. We expect that after successful recombination E. coli cells will glow in different colors located at different positions within the cell.
General goal
The development of novel proteins and the Improvement of existing ones by genetic engineering has a tremendous impact on our daily lives. We all benefit from advances in this field be it for medical, industrial or environmental applications. The central goal of our project is the random generation of novel protein variants. The Marburg_SYNMIKRO team 2012 has created ˈThe Recombinatorˈ: an intelligent Genetically Engineered Slot Machine (iGESM). "The Recombinator" can be used to produce large numbers of novel proteins by combinatorial fusion of functional domains.
Combinatorics
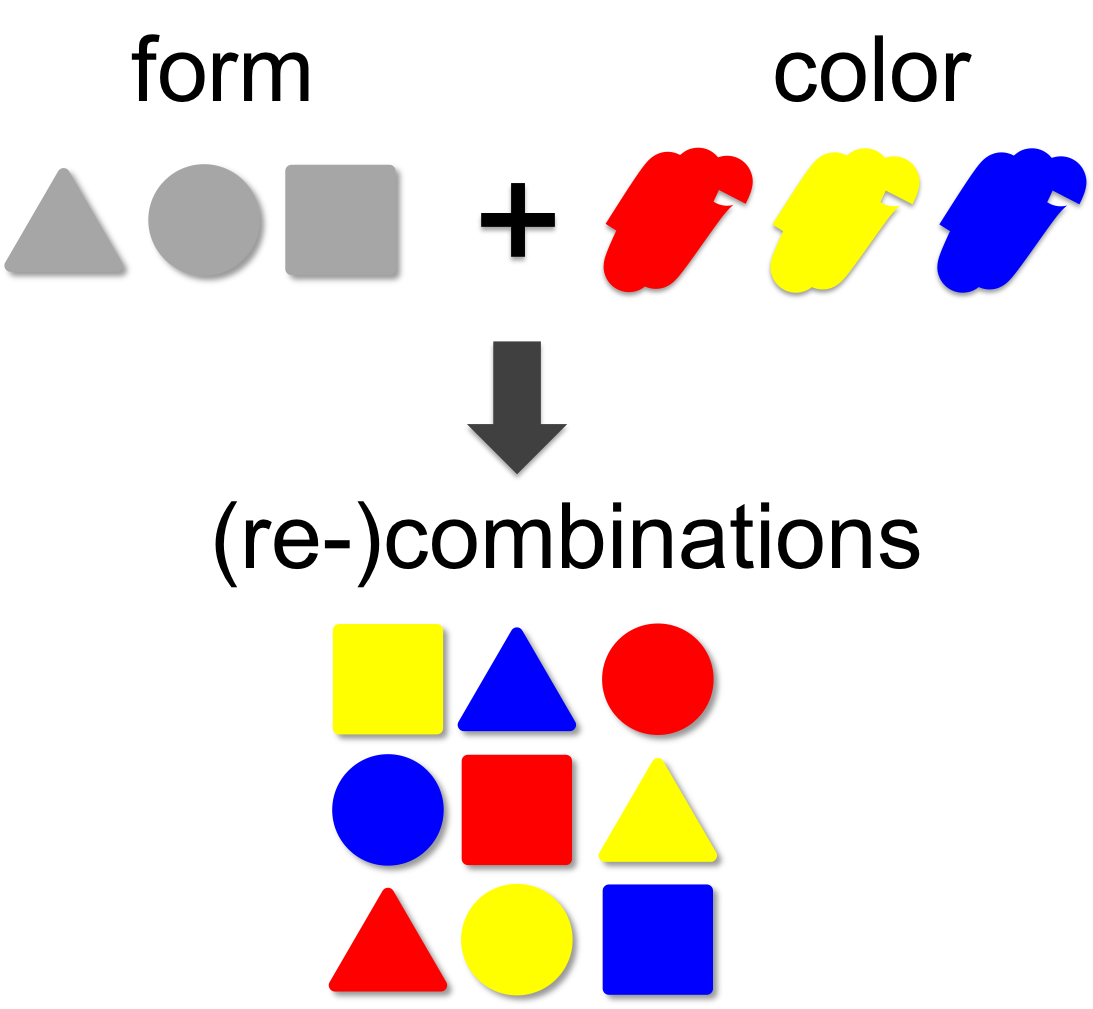
In mathematical theory the number of possible combinations result from multiplication of elements. In our example (left) we combine three forms (triangle, square and circle) with three colors (red, blue and yellow). This results in 3 x 3 = 9 different combinations. A practical example for the large number of combinatorial permutations can be seen on our team page, where you can generate novel team members by recombination.
Recombination in the vertebrate adaptive immune system
Our system for the generation of novel fusion proteins in vivo was inspired by the generation of antibodies via VDJ-recombination. The huge diversity of antibodies is generated by recombination of different segments that are combined into mature antibody chains. This occurs in the B-cells of the immune system and is essential for the recognition of specific antigens.
Antibodies consist of two heavy and two light chains which are covalently connected by disulfide bonds. Both, the heavy and the light chain can be subdivided into constant and variable regions. The variable parts confer the ability to recognize one distinct antigen. Our immune system is able to recognize billions of different antigens from bacteria, viruses and other microorganisms. This raises the question how this large diversity is generated during development.
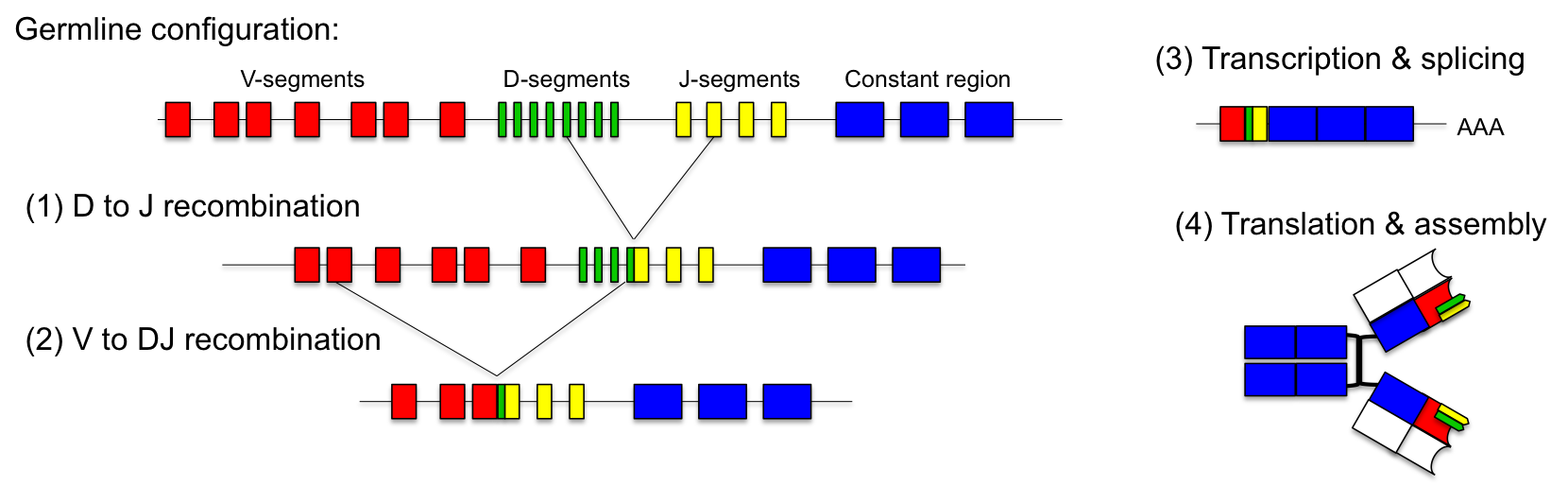
The solution to this problem is the combinatorial design of the variable regions of the antibodies. On the genome level each locus of the light chains consists of up to 40 V-segments and 5 J-segments. The heavy chain consists of 50 V-, 27 D- and 6 J-segments. During maturation of the immune system different antibodies are produced by random fusion of these segments.This process is called VDJ-recombination and is catalyzed by two specific DNA recombinases (RAG-1 and RAG-2).
For the light chain one V-segment gets fused to one of the J-segments by recombination. The production of the variable heavy chain occurs in two steps. Initially, one of the J-segments is fused to the D-segment. Then, the combined DJ-sequence is added to one of the V-segments. The recombined antibody gene is transcribed together with the constant part of the antibody, which is added as exon by splicing. Since each antibody is generated by combinatorial fusion of one of 40 V-segments, 5 J-segments for the light chain and one of 50 V-, 27 D- and 6 J-segments for the heavy chain, the number of possible combinations amounts to 1.6 x 106. Furthermore, the recombination process introduces additional mutations which increase the possible rearrangements to more than 1010.
Site-specific genetic recombination
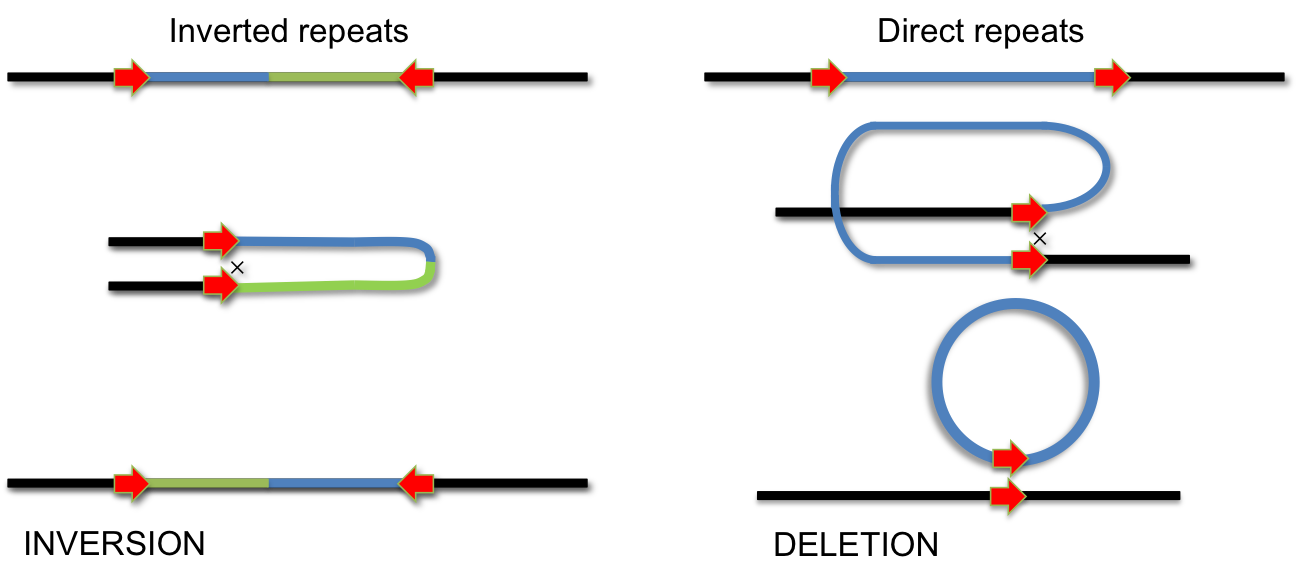
During evolution several systems have invented site-specific DNA recombination systems. Specialized enzymes recognize short DNA elements which are flanking a segment of DNA. Recombination between these short elements results in INVERSION of the DNA segment if the elements are arranged as inverted repeats. If they form direct repeats the intervening segment is removed by recombination (DELETION). Site-specific recombination is used for integration of phage lambda, phase variation of pathogenic Salmonella strains and during VDJ-recombination during antibody development.
Phage Mu
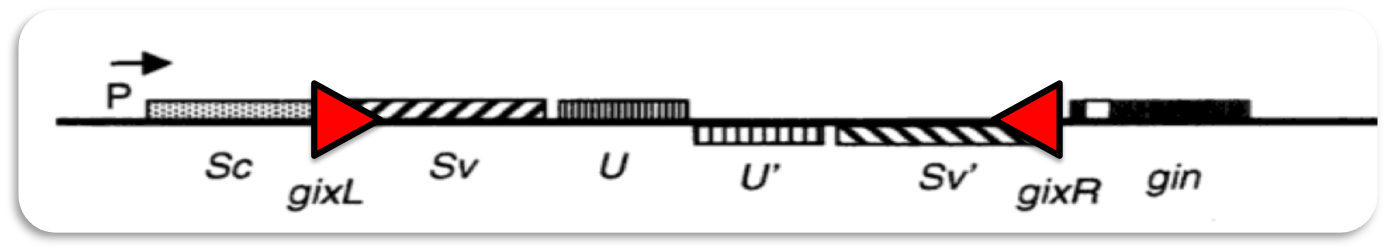
The bacteriophage Mu is a temperate virus which has an unusual lifestyle. It propagates by replicative transposition. Upon entry in its host it integrates into the host genome at a random position. This results in mutations hence the name ("mutator phage")Phage Mu can infect different types of bacteria. To achieve recognition of different hosts the phage genome contains an invertible region of DNA (G-segment). Dependent on the orientation of the G-segment, G(+) or G(-), different lipopolysaccharides are recognized on the surface of bacterial hosts.
Gin recombination
Inversion of the G-segment is catalyzed by the site-specific DNA recombinase Gin which is encoded on the phage genome.
Enhancer
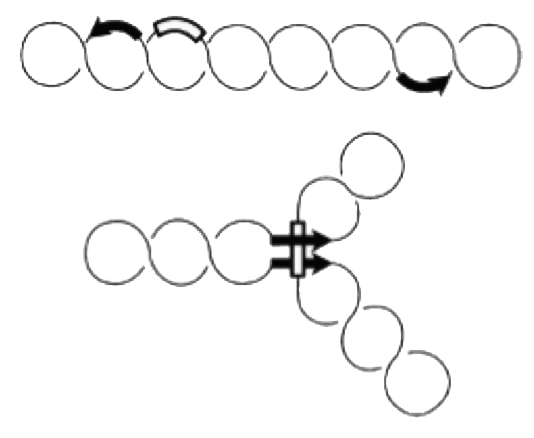
Site-specific recombination catalyzed by Gin recombinase requires the formation of a synaptonemal complex with aligned recombination sites (below). The effective formation of this complex depends on the presence of a recombinational enhancer which in our case is located within the deleted DNA segment.
Construct
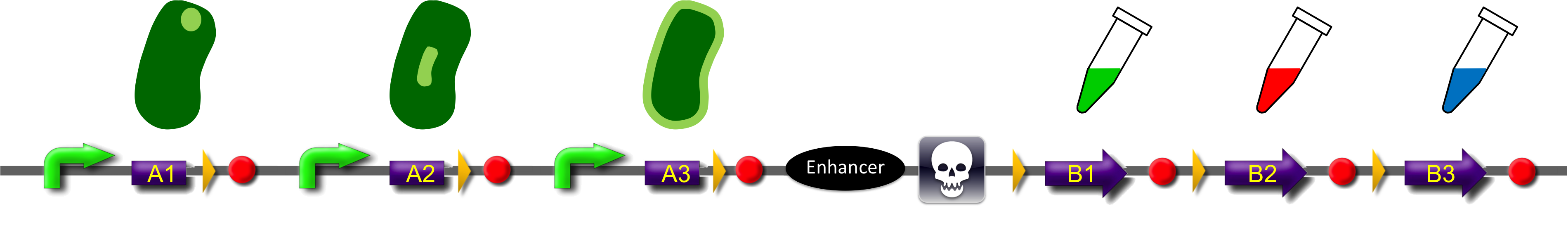
ˈThe Recombinatorˈ is able to recombine diverse protein domainsresulting in expression of chimeric proteins. The system is based on the site-specific Gin recombination system of bacteriophage Mu. Efficient recombination between two gix sites (yellow triangles) requires the presence of an enhancer element (Klippel et al., 1993). To demonstrate the proof-of-principle of our system we placed the enhancer element between bacterial intracellular localization domains (the A-modules) and the fluorescent proteins GFP, CFP and mRFP (B-modules). Due to this design recombination terminates after deletion of the central segment. This guarantees that our system generates a random fusion of one A-module with one B module.
Each localization domain is preceded by a promoter and ribosomal binding site.
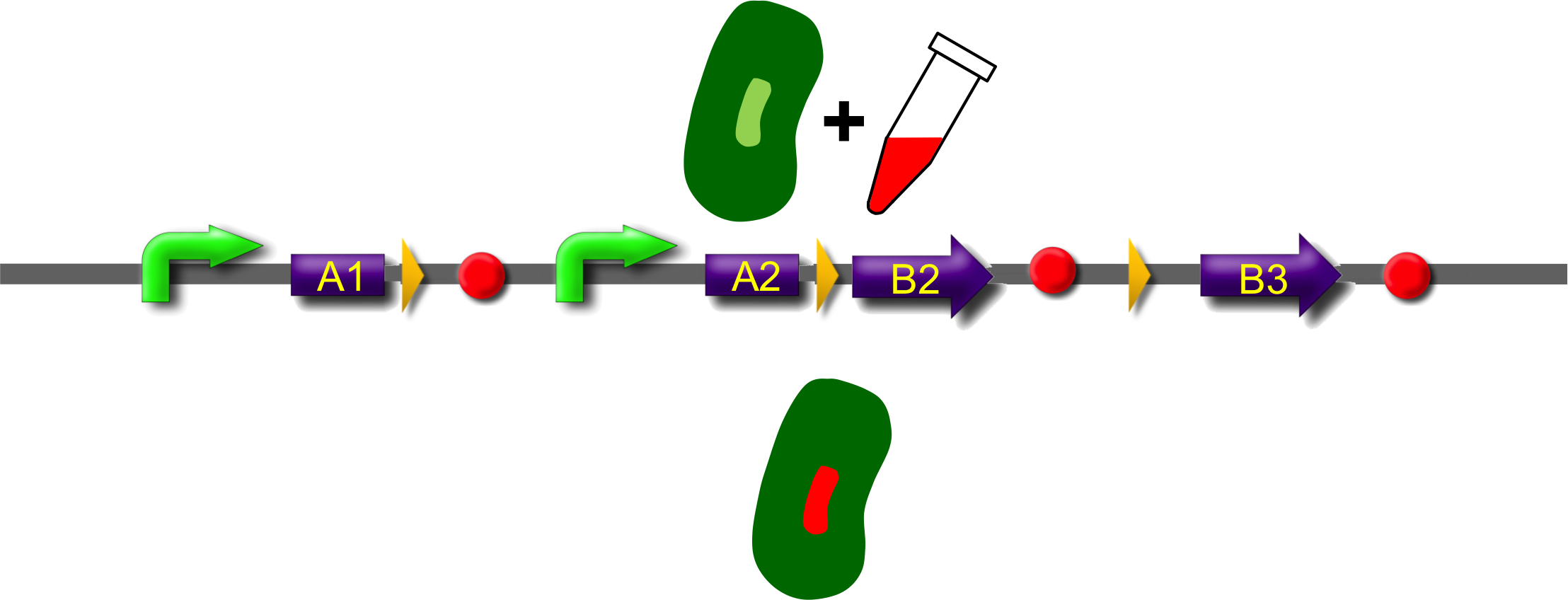
These A-modules are taken from HU, GroES and amidase c. Each of them has the ability to site the new engineered protein at a different area in the cell. For our selected domains this are the cell pole, the periplasm and the nucleoid (Bernhardt et al., 2004; Wery et al., 2001; Li et al., 2012). Downstream of the enhancer we placed . In addition to the A- and B-modules our system requires further features for a successful recombination. First of all, we had to mark the recombination sites with the gix-sites. Thus we added a gix-site after every A-module and in front of each B-module. By this setting a single A-module gets fused to one B-module by recombination. Moreover we put a constant promoter ahead of any A-modules for their transcription. For the termination of the transcription a terminator goes along with each B-module and gix-site of the A-modules. To determine the starting point of the translation a ribosomal binding site (RBS) is placed between the promoter and the A-modules. To receive only cells in which recombination of our construct has taken place we used sac b as a suicide gene for selection. Because we wanted to insure that sac b is deleted after recombination it was set downstream of the enhancer. Finally, we made use of an extra plasmid for the Gin-recombinase with an inducible promoter. The assembly of all stated components led to the creation of ˈThe Recombinatorˈ which works like in the following : By inducing Gin, the recombinase can shuffle one A-module to one B-module at the gix-sites randomly. Parts located upstream of the gix-site from the B-module and downstream of the gix-site from the A-module are deleted including the enhancer and sac b. For instance: If A2 is recombinated to B2 then A3, the enhacer, sac b and B1 are removed from the construct and a novel protein A2/B2 arises. This protein is supposed to be found at the cell pole and to fluoresce red. The usage of three A- and B-modules results in nine possible combinations of the modules but can be scaled up by the utilization of further domains. By imitating a kind of evolutional process, which serves in the creation of proteins, there are certain advantages of our system. Modified enzymes can reveal a higher specificity to their substrates or increase their activities by a extended range of substrates. Moreover proteins can acquire improved functions like alternative partners of interaction or a changed stability. Furthermore our system does not depend on rational design that requires the three dimensional structure of the proteins. Besides industrial applications such as the optimization of cellulases, lipases, amylases or the production of synthetic chemicals in novel metabolic pathways the development and improvement of pharmaceuticals and antibiotics attracts the attention. This is due to their causation of side effects and complications which limit their usage. The discovery of potential pharmaceuticals can be overcome by the generation of combinatorial libaries or phage display which can be done by our construct.
tmRNA
Because of the upstream localized promoter of the A-modules they can be transcribed in mRNA which is recognized by ribosomes. After the production of a polypeptide the ribosomes are released from the mRNA by a stop codon at the 3ˈ end of the translated mRNA. Due to the fact of missing stop codons in the A-modules the ribosomes are stalled at the mRNA. To rescue stalled ribosomes bacteria own a chimeric RNA molecule called tmRNA (transfer-messenger-RNA). This RNA consists of a tRNAAla at the 3ˈ end followed by ten codons that encode for a tag and a stop codon. The tRNAAla is charged with alanine and binds EF-Tu-GTP. If a ribosome is now stalled by a missing stop codon of the A-module the tmRNA binds to the A site of a ribosome and a transpeptidation takes place. At the end of the tmRNA the stop codon leads to the release of the ribosome. This results in the recycling of the ribosomes and in the degradation of the tagged protein by proteases.
Localization domains
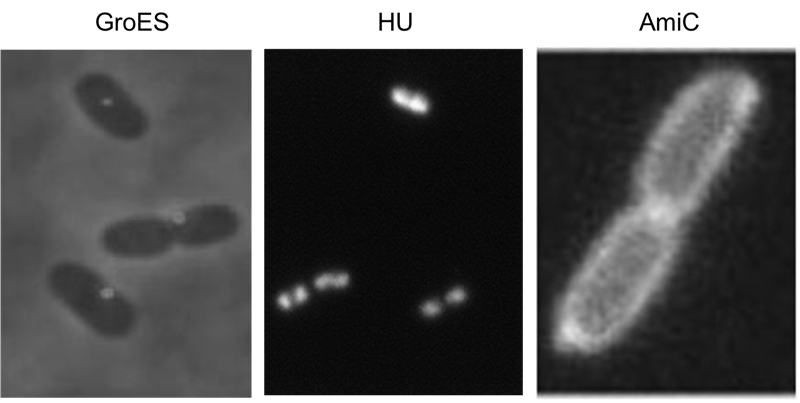
We have screened the literature and identified several bacterial proteins with particular localization within the cell. We decided to use GroES for localization at the cell poles, the small DNA binding protein HU to mark the nucleoid and the amidase AmiC, which is targeted to the periplasm. Depicted are micrographs from the articles by Li et al., 2012, Wery et al., 2001 and Bernhard et al., 2004.
For the examination of a successful recombination we fused a localization domain (A-module) to a fluorescent domain (B-module). However the chosen domains had to comply certain conditions. First of all the domains of the A- and B-modules should possess the ability to yield in a functional protein after their fusion. Moreover the domains had to be distinguishable to demonstrate diverse combinations of the domains. To achieve entirely active proteins we removed any stop codon at the end of the A-modules and tried to avoid their formation at the fusions agency. The applied A-modules should be kept apart due to their localizations inside the cells. So we decided to utilize the domains of HU, GroES and amidase c. HU belongs to a group of ˈhistone-likeˈ small DNA-binding protein and has its function in the organization of the genomic DNA. It is present in Escherichia coli, Eubacteria and homologs of it can be found in several species of bacteria and chloroplasts (Ram et al., 2008; Karcher et al., 2009). So B-modules fused with this domain should be recognized in the area of the nucleoid. Furthermore we used the chaperonin GroES to place fusion proteins at the cell pole. GroES interacts in a complex with GroEL and provides protein folding in the presents of ATP (Li et al., 2012). The third domain was taken from amidase c. Amidase c hydrolyses the cell wall during the cell division and can be localized at the periplasm (Bernhardt et al., 2004). By detection of the generated proteins at different areas of the cell we can ensure that all A-modules are able to fuse with the B-modules.
To assure the same for the B-modules they encode for different fluorescent proteins. In the construct gfp, cfp and mRfp permit guarantee for different combinations of the B-modules with the A-modules.
Fluorescent proteins
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
The Experiments
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Results
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Outlook
pipapo
References
Bernhard TG and de Boer PA (2004) Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol 52:1255-1269.
Klippel A, Kanaar R, Kahmann R and Cozzarelli NR (1993) Analysis of strand exchange and DNA binding of enhancer-independent Gin recombinase mutants. EMBO J 12:1047-1057.
Li G and Young KD (2012) Isolation and identification of new inner membrane-associated proteins that localize to cell poles in Escherichia coli. Mol Microbiol 84:276–295.
Wery M, Woldringh CL and Rouviere-Yaniv J (2001) HU-GFP and DAPI co-localize on the Escherichia coli nucleoid. Biochimie 83:193-200.
 "
"