Team:Kyoto/Project
From 2012.igem.org
Contents |
General Introduction
A technology blooming flowers on demand will bring a lot of fruits in the field of agriculture. We try to achieve this goal by our Flower Fairy E.coli, which can produce Florigen. Florigen is a kind of plant hormone, which is a peptide composed of one hundred and several tens amino acid. This small peptide hormone activates the transcription of some genes inducing blooming and is effective at low dose. The main purpose of our project is to make Florigen in E.coli and introduce this hormone into Plants!
We also execute sub goal. We study how a gene express when many promoters are inserted into a gene.
In order to achieve our goal, we devided our team into 3 groups――Florigen, Secretion, and Golden Gate Assembly.
Florigen
Abstract
FLOWERING LOCUS T (Florigen, FT protein) is made in the leaves of plants and leads flower formation. We will make "Flower Fairy E.coli" that produce and introduce florigen into plant cells, and makes flowers bloom. Flower Fairy E.coli makes FLOWERING LOCUS T (Florigen), secretes florigen through TAT secretion system, and introduces florigen into plant cell using R9 peptide.
This florigen group tries to confirm that E.coli can make florigen that acts correctly and that protein with R9 peptide can penetrate cell wall and membrane.
Introduction
Motive for Flower Fairy E.coli
Flower fairies live only in fairy tales, but it would be very lovely if they actually existed. Not only they would entertain us, but also their talent for blooming flowers at will would be greatly profitable for us in many aspects, such as agriculture.So we set our project to realize them in real world by synthetic biology.
In order to obtain our goal, it is essential to utilize materials which can control and trigger flowering freely. We noticed that a plant hormone, Florigen, can satisfy above goal.
Even though various studies have been done to realize dream, little is known about florigen. It is a plant hormone which was suggested and named seventy years ago. Since then, many studies have revealed its function more and more. We would tell you some of studies below.
Florigen is a 20kDa of small protein composed of 175 amino acids which are encoded by the FLOWERING LOCUS T (FT) gene. The expression of the FT gene is promoted inside the leaf phloem companion cells by several specific conditions such as long photoperiods, vermalisation, age and so on. After transcripted, FT mRNA is translated into a protein and then transported to the shoot apex thorough sieve tubes. In the shoot apex, FT is combined with FD, which works as a transcriptional factor, and activates floral meristem identity genes such as APETALA 1 (AP1), FRUITFUL (FUL), and LEAFY (LFY). These genes play a significant role in the transformation of the form of a shoot meristem, converting it into a flower bud.
Considering the effect of FT on flowering as mentioned above, we can say that E. coli with a capacity of producing FT are able to bloom flowers, as if they were flower fairies!
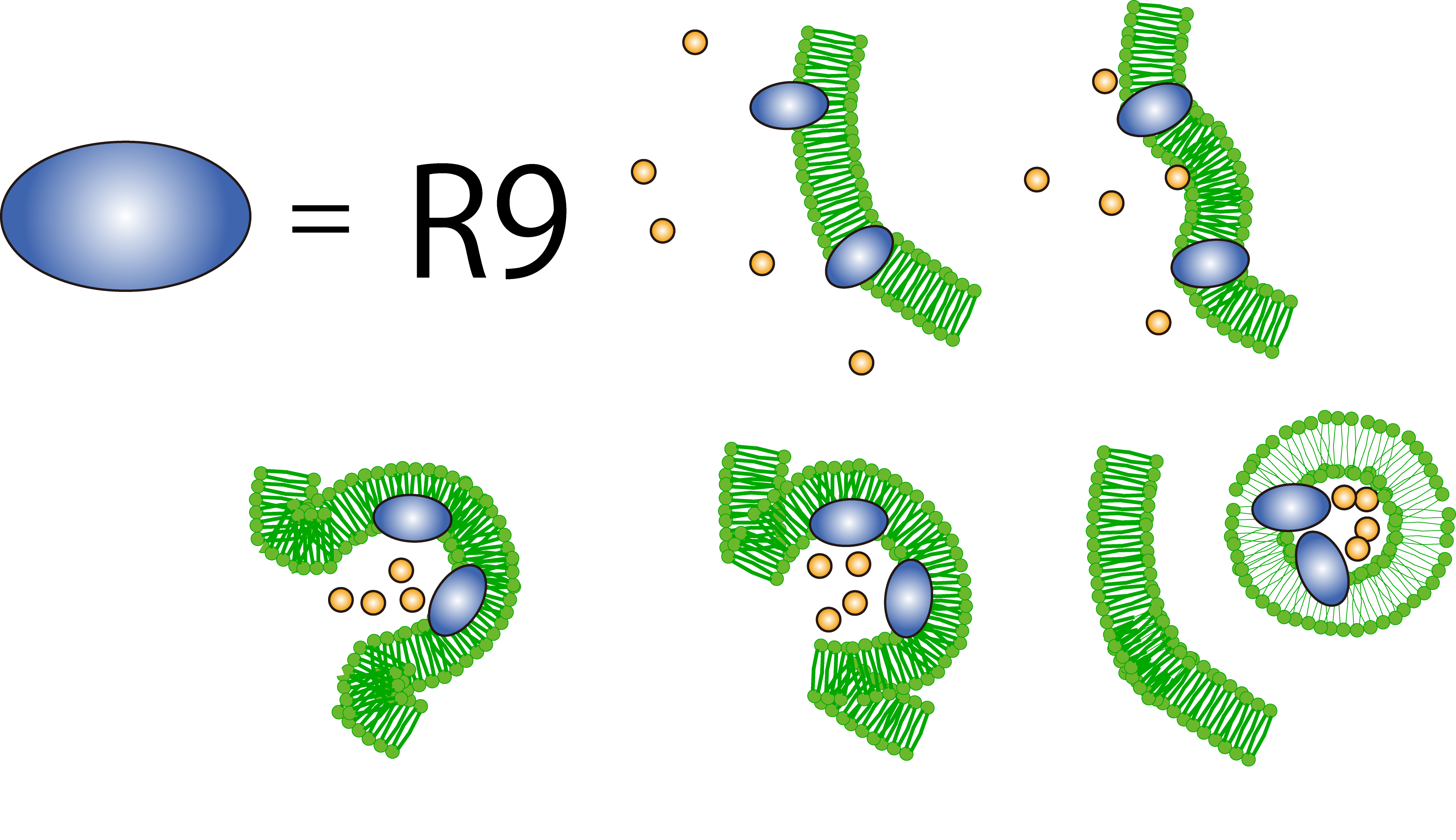
What is R9 peptide?
To realize flower fairy E. coli, FT secreted by E. coli must be absorbed in plant cells. We used polyarginine, a type of cell penetrating peptide (CPP), in order to transport FT protein into plant cells. The type is R9 peptide, which comprises a sequence conjugated to nine arginine residues.
Polyarginine peptide is thought to act
on a cell membrane and cause a specific form of endocytosis, that is, macropinocytosis. Macropinocytosis is not
caused by an invagination of the cell membrane, but by the growth on the actin mambrane from protrusions into
vesicles called macropinosomes, and no receptors are necessary for the process. It is reported that plants use
CPP to transport biomolecules such as proteins inside the cell, in spite of their cell walls.Method
1. Expression of FT in E.coli
Construction
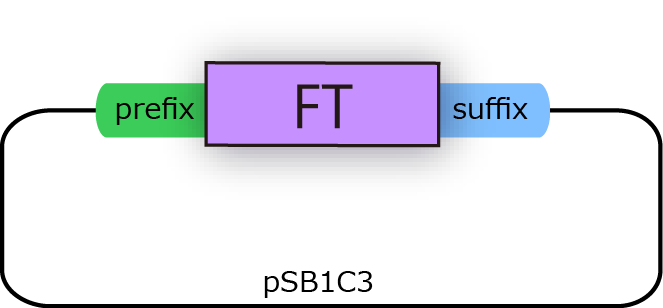
FT gene has 2 BioBrick restriction enzyme sites, EcoR1 and Pst1 (fig.3A.) So we did mutation to delete the sites
before we start construction.
We successfully cloned FT gene into BioBrick plasmid backbone, pSB1C3 with prefix and suffix (fig.3B.)
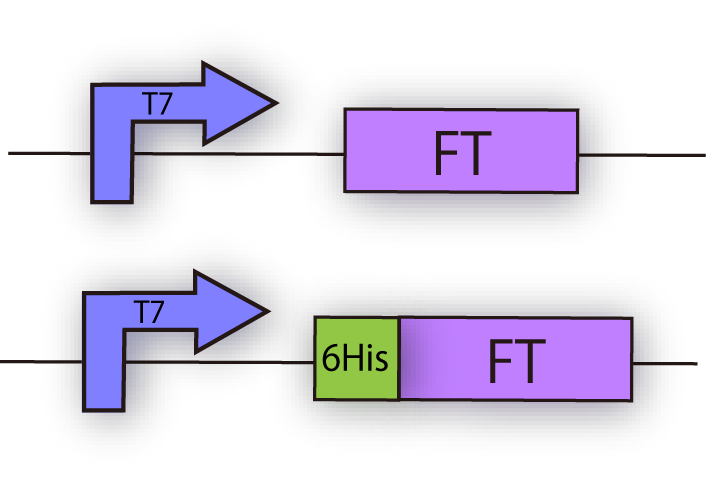
We constructed the following plasmid to check the expression of FT by Western blotting, using FT specific
antibody. FT and 6x his tagged FT are regulated by T7 promoter, BBa_I719005 and strong RBS, BBa_B0034
(fig.3C.)
Western blotting
1. Pre-experiment
First, we checked the specificity of anti GFP monoclonal antibody.
We characterized the existing GFP generator parts, [http://partsregistry.org/Part:BBa_I746915
BBa_I746915].
The parts is consist of T7 promoter 6-his tagged superfolder GFP.
Cells were precultured overnight and diluted into fresh SOC medium. IPTG was added when OD600 was approx.
0.7, then cells were incubated for 4h at 37°C. 100µL of culture was used for SDS-PAGE.
2. R9 peptide fusion GFP
Next, using following construction, we tried to check the expression of R9 peptide fusion GFP. Cells were precultured overnight and diluted into fresh SOC medium. IPTG was added when OD600 was approx.
0.7, then cells were incubated for 4h at 37°C. 100µL of culture was used for SDS-PAGE.
3. GFP imaging assay for characterizing R9 peptide
Following plasmid was constructed to check the function of R9 peptide.
4. RT-PCR assay for characterizing FT
[http://partsregistry.org/Part:BBa_I746915
BBa_I746915].
The parts is consist of T7 promoter 6-his tagged superfolder GFP.
Cells were precultured overnight and diluted into fresh SOC medium. IPTG was added when OD600 was approx.
0.7, then cells were incubated for 4h at 37°C. 100µL of culture was used for SDS-PAGE.
Verification of R9 system
1.skin cuticules of plants leaves with the use of the head of a pencile.
2.soak them in a solutiuon of GFP and R9 or only GFP 20 ul per 1 sample.
incubate for 5 minutes
3.wash them by PBS 60ul per 1 sample
4.Hoechst dyeing
5.observe them by confocal microscope.
Result
We have to go through four steps for purpose of obtaining our goal-Flowering Fairy E.coli-
The four steps are composed of “EXPRESSION”,”SECRETION”,”PENETRATION”, and ”ACTIVATION”
1.EXPRESSION
On the first step; EXPRESSION, E.coli produce florigen inside of the cells
When we got FT gene, we had a difficulty in constructing iGEM parts. The problem is that FT sequence had two cleavage sites of iGEM restriction enzymes. In order to eliminate cleavage sites of iGEM restriction enzymes, we performed Inverse PCR of plasmids with two kinds of primer which have mutation.
As a result, we could get mutated plasmids, which are not cleaved by iGEM restriction enzymes. In this way, we made FT gene available.
Previous to experiment, whether FT gene is expressed correctly in E.coli had arouse criticism in our team. This is because, under natural environment, FT gene is expressed and functions in plant cells, which are eukaryote. However, E.coli is prokaryote, so no one knows that FT gene which E.coli made could function.
We transformed E.coli and made them express our FT gene. In order to confirm the expression of FT protein, we performed western blotting and checked the place of the FT protein band.
1. Pre-experiment
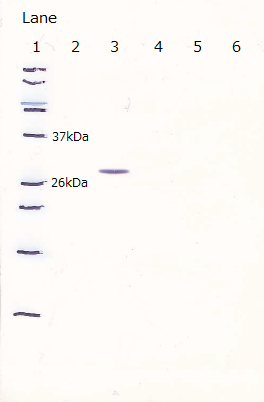
Following figure shows the result of western blotting of [http://partsregistry.org/Part:BBa_I746915
BBa_I746915].
Unfortunately, we used inappropriate molecular marker and could'nt confirm the molecular weights of samples.
Lane1 : IPTG 0mM, sample 10µL
Lane2 : IPTG 1mM, sample 10µL
Lane3 : IPTG 0mM, sample 5µL
Lane4 : IPTG 1mM, sample 5µL
Lane5 : IPTG 0mM, sample 2µL
Lane6 : IPTG 1mM, sample 2µL
2. R9 peptide fusion GFP
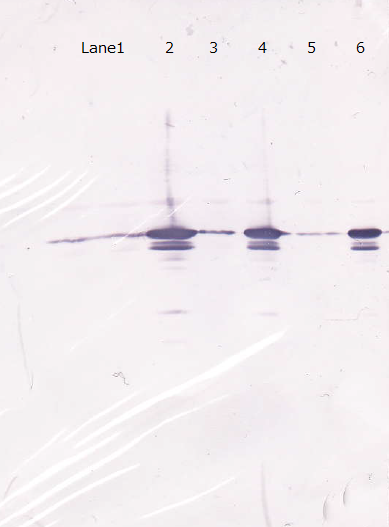
After the 4h of IPTG induction, we noticed that E.coli expressing R9::GFP were growing poorly.
Moreover, we couldn't get any bands of R9::GFP, as shown in the following figure.
Lane1 : Molecular marker
Lane2 : GFP([http://partsregistry.org/Part:BBa_I746915 BBa_I746915]) IPTG 0mM, sample 10µL
Lane3 : GFP([http://partsregistry.org/Part:BBa_I746915 BBa_I746915]) IPTG 1mM, sample 10µL
Lane4 : R9::GFP IPTG 0mM, sample 10µL
Lane5 : R9::GFP IPTG 1mM, sample 10µL
3. Separating R9 peptide and GFP
When R9 and GFP were conected, they didn't work normally. By way of experiment, we separated R9::GFP into
two segments and soaked plant cells into a sollution of them.
[[]]
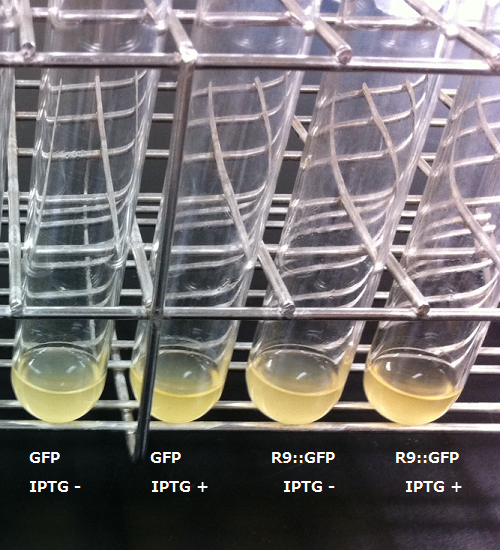
The control on the left was soaked in only GFP, and on the right-hand side the sample was soaked in GFP and
R9. These two pictures show the action of R9 peptide. R9 peptide kept GFP in or around plant cells. This figure
strongly suggests that R9 peptide works successfully and penetrates cell membrane with GFP.
4. Verification of FT Function
Finaly, we verified whether FT normally worked in plant cells. Probably R9 penetrates cell membranes with proteins, so we injectd a sollution of FT and R9 into plant cells derived from Arabidopsis thaliana. FT protein activates various other proteins. We checked the change of activity of them by RT-PCR.
[[]]
This is the result of RT-PCR. The left is control. It is
Discussion
Reference
[1]Microsugar Chang et al. (2005)"Cellular internalization of fluorescent proteins via arginine-rich intracellular
delivery peptide in plant cells" Plant Cell Physiol, 46(3), 482–488
[2]Paula Teper-Bamnolker and Alon Samach1 (2005) "The flowering integrator FT regulates SEPALLATA3 and
FRUITFULL accumulation in Arabidopsis leaves" The Plant Cell, 17, 2661–2675
[3]Philip A. Wigge et al. "Integration of spatial and temporal information during floral induction in Arabidopsis
[4]Sara Trabulo et al.(2010). "Cell-penetrating peptides—mechanisms of cellular uptake and generation of delivery
systems" Pharmaceuticals, 3, 961-993
Secretion Group
Abstruct
Even though our fairies can produce FT protein, there remains a big issue: how they can transport proteins to the outside of the cells? To make it possible, we made Tat cassette and kil protein inducer. This cassette lets E.coli carry proteins with torA signal via Tat protein transportation pathway from cytoplasm to periplasm and Kil protein encourage proteins to move from periplasm to surroundings. We made this protein secretion system and visualized and confirmed its functioning by using GFP.
Introduction
We need secretion system without cell-death
In previous iGEM competition, some kinds of parts and devices for protein translocation were already developed. One of the most widely used parts is lysis cassette(このパーツのページのリンク). This part causes cell lysis and, as a result, makes E.coli scatter materials it includes. This style is, unfortunately, not suitable for our project because of the possibility of accidental all-death. Generally speaking, the concentration of E.coli on flower is not high so that it can’t be ignored that the possibility of occurring all cell-death. Once all our Fairies disappears, flower wouldn’t bloom. In addition to that, lysising E.coli is NOT CUTE.
We tried to seek for a ideal secretion systems, which is not harmful and dangerous for both of our fairies and other livings. Finally, We focus on Twin argenine translocation pathway (Tat pathway) of E.coli. It is not pathogenic and carries various kinds of proteins with torA signal(Tat班古い方のレビュー)
What is TAT Secretion pathway
The Twin Arginate Translocation pathway(TAT) is secretion system E.coli originally have. This system can carry proteins that have torA signal anino acid sequences at N terminal. TatA, TatB, TatC and TatD compose Tat complex on inner membrane. Tat complex recognizes torA signal peptide and then it transports protein (with torA) from cytoplasm to periplasm. In addition, protein that has passed through TAT pathway cut off the torA signal. Proteins which are secreted by this system have no tag that obstruct the activation of the protein.
Our Tat cassette and kil inducer
TatABCD composes pathway from cytoplasm to periplasm. Kil makes holes on outer membrane and we expect that protein goes through this holes. Needless to say, function of outer membrane as membrane is essential for E.coli to survive. In other words, overexpression of Kil causes cell death. In this reason, we must find suitable amount of expression.
Detail of Our Secretion System
Our wonderful secretion system is constructed by tatABCD, Kil and another gene. Another gene is PspA (phage-shock protein A) gene. E.coli has it originally and this gene is expressed when their inner membrane is dameged. PspA meintains H+ concentration gradient between periplasm and cytoplasm and membrane potential.
Our secretion system makes many holes on inner and outer membranes. In other words, E.coli which has our secretion system is under the membrane stress conditions. But by introducing pspA into our Flower Fairy E.coli, the E.coli come to be able to maintain the vitality, though they have many holes on the membrane.
Method
Observation with confocal microscope
- PFA
- MilliQ
- Hoechst
- Slide glass
- Gather fungus body by centrifugal separation
- Add PFA to each microtubes containing fungus body and resuspend them
Results
Construction
Tat secretion cassette with constitutive promoter(BBa_K797004)
This cassette allow E.coli to secrete proteins with torA signal. Wild type Tat protein secretion system is too week so that Kyoto 2012 construct Tat cassette to reinforce the ability of transportation of Tat system.
This parts include tatA,B,C protein coding region and pspA (phage shock protein A). Tat A,B,C protein is the main
component of Tat complex where proteins with torA signal go through and pspA can encourage protein secretion via Tat system. Kyoto 2012 suggest iGEMers with this new way of secretion and provide them with this cassette regulated by constitutive promoter.
We checked the sequence of tatABCD(BBa_K797000) and the sequence of pspA(BBa_K797001) individually, and then, we made TAT construction composed of constitutive promoter(BBa_J23107), tatABCD(BBa_K797000),pspA(BBa_K797001) and double terminator(BBa_B0015). This TAT secretion cassette is too long device to sequence, so that we performed electrophoresis of this cassette and confirmed the length of our parts.
Considering that the sequences of tatABCD and pspA are correct,and the length of TAT secretion casssette is correct, we declare that this construction of TAT secretion cassette has been completed.
Discussion
References
[1]Tracy Palmer and Ben C. Berks "The twin-arginine translocation (Tat) protein export pathway"
[2]J. H. Choi. S. Y. Lee "Secretory and extracellular production of recombinant proteins using Escherichia
coli"
[3]G. Miksch · E. Fiedler · P. Dobrowolski · K. Friehs "The kil gene of the ColE1 plasmid of Escherichia coli
controlled by a growth-phase-dependent promoter mediates the secretion of a heterologous periplasmic protein
during the stationary phase"
[4]Brad A. Seibel* and Patrick J. Walsh "Trimethylamine oxide accumulation in marine animals: relationship to
acylglycerol storage"
Future Works
We noticed only flowering and florigen in this time but there are many many other plant hormones. We made translocation pathway from E.coli into plant cells, so we will be able to introduce plant hormones into plant cells if E.coli can make them. It means we can control plant growth in any stage through genetically engineered E.coli. In the future that is not so far, we will be able to meddle in plants' growth――germinating, elongation, flowering, and fructification. We human will finally accomplish a technology that control plants perfectly.
Moreover, R9 peptide functions not only plant cell. R9 peptide works on animal cell similarly. It means that we found a pathway into any kinds of cells. R9 peptide tag enables us to introduce proteins into any cells, so we will be able to controll all living cells using this technology.
Golden Gate Assembly
Introduction
BioBrick is useful for us because we can look for required BioBrick parts from its registory and recombine genes easily. If we want to introduce many parts into one plasmid, however, we have to repeat the process; restrict enzyme digestion and ligation. It takes us too much time and sometimes we lose enough time for other experiments.
We want to reduce the time required for the recombination of genes and get enough time for verification of the expression and the effect of genes.
Golden Gate Assembly is the one of the ways to make it possible.
What's Golden Gate Assembly
Golden Gate Assembly is a method which enables us introduce plural
gene segments into one plasmid all at once. We don't need to digest
gene by restrict enzyme before ligation because restrict enzyme digestion
and ligation are compleated by just one PCR.
Golden Gate Assembly use the feature of restrict enzyme "BsaI".
BsaI recognizes the sequence "GGTCTC" and cut DNA like the figure.
And BsaI activity is independent of the sequences of the downstream of the
recognition site.
 "
"