Team:Bielefeld-Germany/Labjournal/week20
From 2012.igem.org
Contents |
Week 20 (09/10 - 09/16/12)
weekly meeting
- We are discussing our lab results. Some experiments have to be repeated, but we are on a good way.
Monday September 10th
- Team Site Directed Mutagenesis:
- Digestion of the six tvel10-plasmids. Three were unmutated and other three hadn't lost the illegal SpeI-restriction-site, but their second fragment was of a smaller size.
- plated three additional tvel-t243g-colonies.
- Team Cellulose Binding Domain:
- Sequencing results arrived: One [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] is completely right!
- Nanodropping plasmids of isolated [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His] showed that the cells did not have a plasmid at all (selection-agar did not work)
- Gradient-PCR with [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] as template and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg]-primers did work just fine (best temperature 61,7°C); Digestion with XbaI and AgeI.
- Gel-Clean-up of [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] and [http://partsregistry.org/Part:BBa_K863121 GFP_His]
- Digestion of [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] with XbaI+AgeI.
- Digestion of [http://partsregistry.org/Part:BBa_K863121 GFP_His] with NgoMIV and PstI.
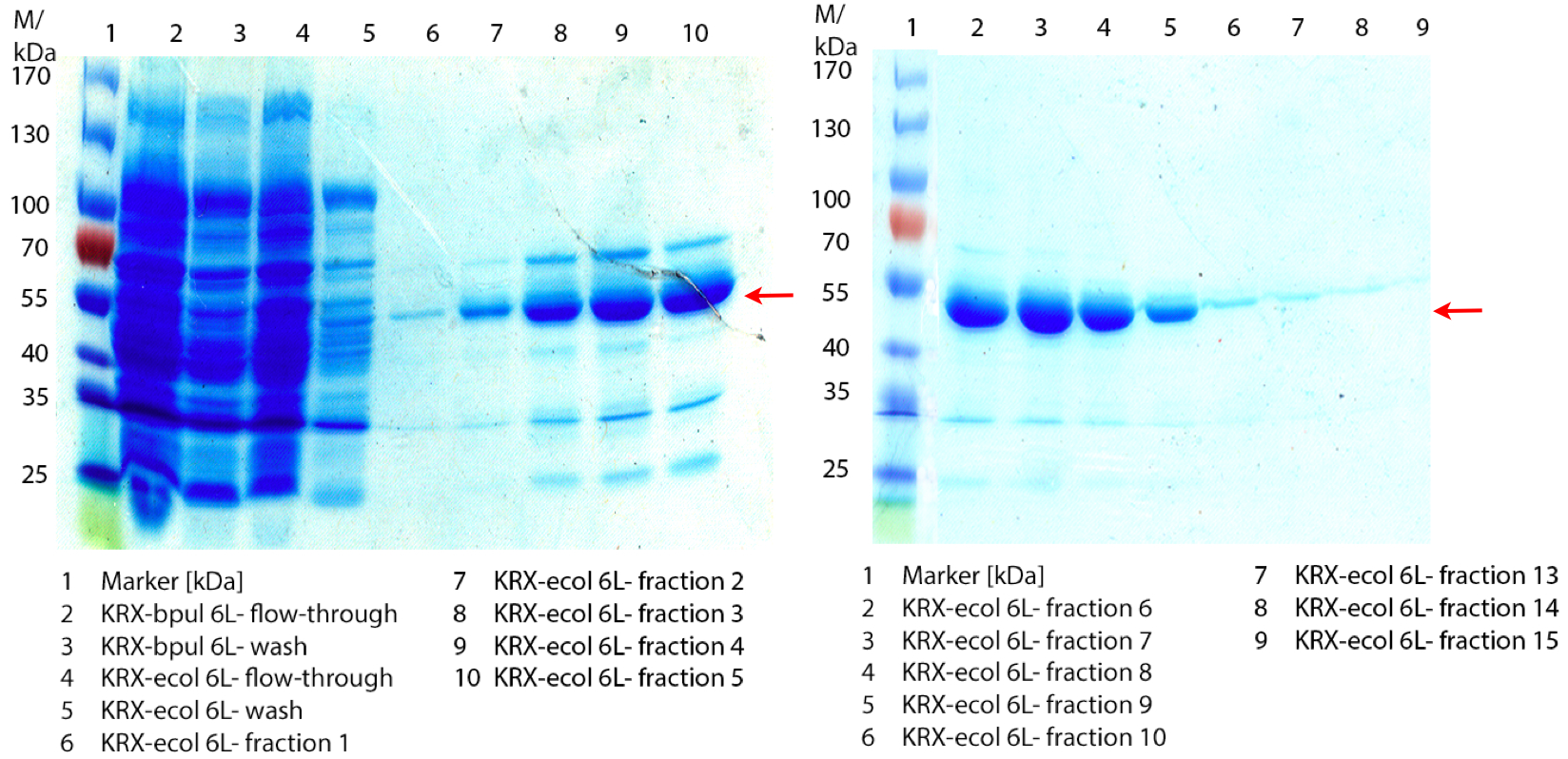
Figure 1: Fermentation from 09/07 of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], 6 L in NFL22, autoinduction medium, 37 °C, pO2 30 % for 12 hours. Purification of the supernatant via Ni-NTA column. The red arrow shows ECOL.
- Team Cultivation & Purification:
- Cell disruption via high-pressure homogenizer and purification via Ni-NTA column were performed for the following samples: 3 L fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (09/09), 6 L fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] (09/07) and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (09/09).
- We made SDS-Pages of purification of ECOL.
Tuesday September 11th
- Team Shuttle Vector:
- Digest of shuttle shuttle vector with PvuII and HindIII as control. Agarose gel looks good.
- Team Fungal and Plant Laccases:
- PCR on tvel5 laccase for cloning in shuttle vector. Digest of shuttle vector and digest of tvel35 with AarI enzyme, ligation and transformation in E. coli KRX cells.
- Team Cellulose Binding Domain:
- Assembly of <partinfo>BBa_K863104</partinfo> and <partinfo>BBa_K863114</partinfo>:
- Assembled [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] and [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] with [http://partsregistry.org/Part:BBa_K863121 GFP_His] and <partinfo>J61101</partinfo> and plated the ligation on AMP-selection-agar (because of the pSB1A2-backbone).
- Restriction of [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] and [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] with EcoRI and PstI.
- Ligation of CBDcex_Freiburg and CBDclos_Freiburg with the pSB1C3-backbone and transformated and plated on selection-agar
- Assembly of <partinfo>BBa_K863104</partinfo> and <partinfo>BBa_K863114</partinfo>:
- Team Cultivation & Purification:
- Made precultures of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] as well as of E. coli KRX.
Wednesday September 12th
- Team Fungal and Plant Laccases:
- Ligation of shuttle vector with tvel5 (digested with AarI).
- Team Site Directed Mutagenesis:
- Plasmid-isolation of the three tvel10-plasmids and digestion with SpeI showed two unmutated plasmids and one with the same wrong restriction-fragments as Monday. There must be a systematical error. pfu-PCR should be done again.
- Team Cellulose Binding Domain:
- There are only few colonies on all selection-agar-dishes, but none is obviously green fluorescing, even with UV-light emission could not be stimulated.
- Plated colonies of [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] to see if they are red or not.
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] showed eight positive colonies to the <partinfo>K863104</partinfo>-insert. Plated the positive colonies for plasmid-isolation.
- Team Cultivation & Purification
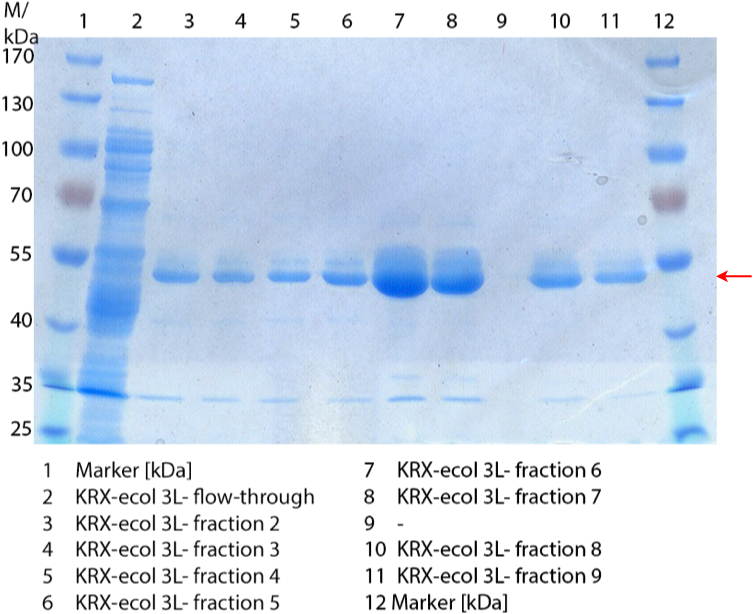
Figure 1: Fermentation from 09/09 of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], 3 L in Infors, autoinduction medium, 37 °C, pO2 30 % for 12 hours
- Cell disruption via sonification and purification via Ni-NTA column were performed for the following samples: 200 mL cultivation of E. coli Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09) behind a constitutive promoter.
- Made SDS-Page of the fermentation from 09/09.
Thursday September 13th
- Team Fungal Laccases and Team Shuttle Vector:
- Bad day for us. We realized that our primers we used for amplification of the laccase genes for cloning in the shuttle vector don't have overhanging ends which were needed for an optimal activity of the enzyme AarI. So we had to order new primers with some bases prior to the recognition sites. But after there is little time left until wiki freeze and the new primers need some time until they are here we started a last trial for cloning this 'wrong' PCR products in the shuttle vector. The idea was to phosphorylate and ligate the ends of the PCR product from tvel5 together. Then the problem with the missing bases at the end would be solved and AarI can digest the end of the PCR product. We digested the shuttle vector with AarI and dephosphorylated it. After ligation we transformed the approaches in E. coli KRX cells.
- Team Cellulose Binding Domain:
- Designed a lot of primers to cope with the expression problem. E.g. inserting a long S3N10-Linker via blunt-end-cloning between the CBD and the GFP, also primers to get rid of the His-tag on the GFP and a last set to easily change the order of CBD and GFP (via assembly but with no linker in between).
- Plasmid-isolation of <partinfo>BBa_K863104</partinfo>-transformation clones
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] colonies. All [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg]-colonies are positive and half of the [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg]-colonies.
- Colony-PCR of CBDclos_F.+GFP with no positive result
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863122 const.GFP_His]: 2 positive (one fluorescend); Plated both positive and one additional fluorescend.
- Team Cultivation & Purification:
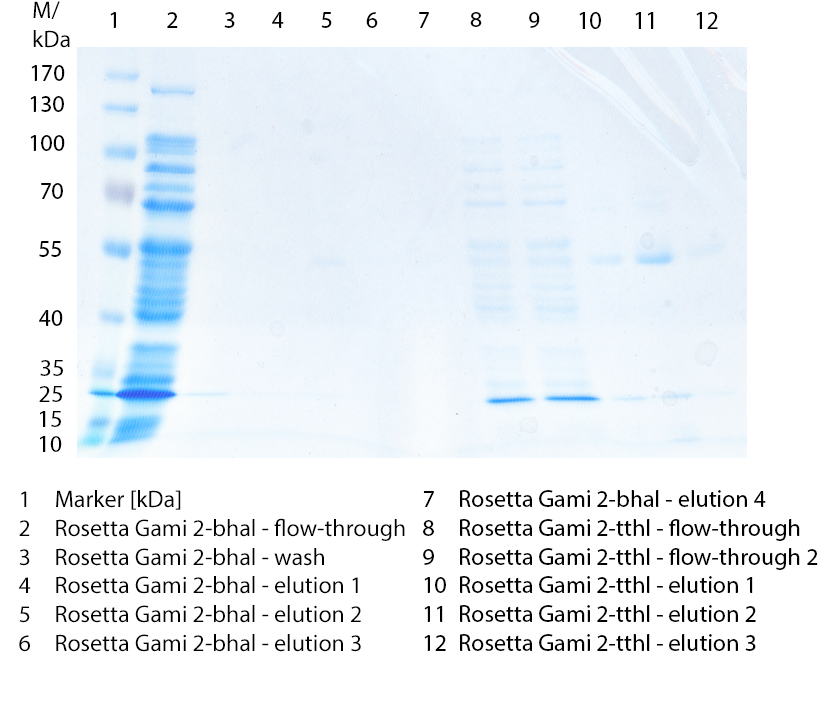
Figure 2: Cultivation from 09/09 of E. coli Rosetta-Gami 2 cells containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012] and <partinfo>BBa_K863022</partinfo>. 200 mL in flasks without baffles, LB medium, 60 µg/mL chloramphenicol and 300 µg/mL ampicillin, 37 °C for 48 hours. Purification of the supernatant via HisTrap column. The red arrow shows TTHL in lane 10-12 (elution 1-3).
- SDS-Pages of the flask cultivation from 09/09 ( E. coli Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo>)
- Fermentation of E. coli KRX without plasmid (fermenter: Infors) and with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (fermenter: Braun Biostat)
- Settings: fermenter: Infors/Braun Biostat, final volume: 3 L, autoinduction medium, 60 µg/mL chloramphenicol added, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NL/m, durance: 12 h.
- Made preculture of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863103 BBa_K863103] (CBD-GFP-His).
- Made preculture of P. pastoris GS115
- Fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000].
- Settings: fermenter: Bioengineering NFL22(7 L), final volume: 6 L, autoinduction medium with 60 µg/mL chloramphenicol added, 37 °C, stirrer increased 2 % if the pO2 got below 30 %, airflow: 5 NL/m, 12 hours.
Friday September 14th
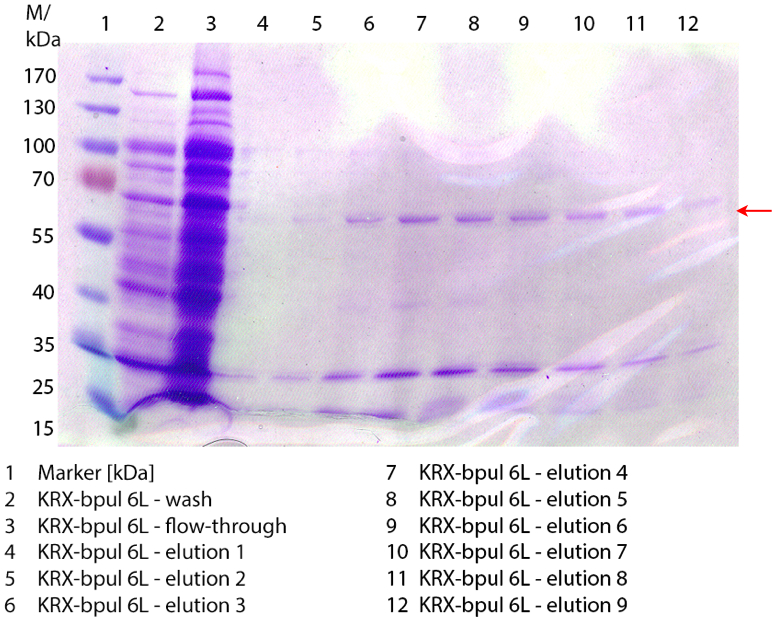
Figure 3: Fermentation from 09/13 of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], 6 L in NFL22, autoinduction medium, 37 °C, pO2 50 % for 12 hours. Purification of the supernatant via Ni-NTA column. The red arrow shows BPUL
- Team Fungal Laccases and Team Shuttle Vector:
- Waiting for colonies from our transformation the day before.
- Team Cellulose Binding Domain:
- Isolated three glowing dishes of KRX with the [http://partsregistry.org/Part:BBa_K863122 const.GFP_His]-plasmid, three with the [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg]-plasmid and [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg]-plasmid.
- Team Cultivation & Purification:
- Cell disruption of fermentation 09/13 via high-pressure homogenizer and purification via Ni-NTA column. Made SDS-Pages of purificated fractions.
- Repeat the preculture of P. pastoris GS115, because of using the wrong media.
- Made preculure of E. coli Rosetta Gami 2 with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012].
Saturday September 15th
- Team Cloning of Bacterial Laccases:
- Digest of <partinfo>BBa_J04450</partinfo> to have pSB1C3 backbone for the last clonings.
- Team Fungal Laccases and Team Shuttle Vector:
- We had some colonies on our LB + CM plates from our transformation of shuttle vector + tvel5. So we did Colony PCR to see if the gene was inserted. But sadly all colony PCRs are negative.
- Team Cellulose Binding Domain:
- KRX culture of [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)-GFP_His] seems to have a green glow.
- Isolation of four pellets of [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)-GFP_His] for us and SDU Denmark.
- [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] digested with SpeI and AgeI and deposphorylated.
- [http://partsregistry.org/Part:BBa_K863121 GFP_His]-PCR-product (gel-clean) digested with SpeI and NgoMIV.
- Ligated [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] with [http://partsregistry.org/Part:BBa_K863121 GFP_His] and plated on select-Agar.
- Restriction-analysis showed that all [http://partsregistry.org/Part:BBa_K863122 const.GFP_His]-plasmids are correct, as are the three [http://partsregistry.org/Part:BBa_K863111 CBDclos] and two of the [http://partsregistry.org/Part:BBa_K863101 CBDcex]
- Collected data to make a protocol for a Cellulose binding assay:
- Avicel: about 11,4 mg protein (CBD) binds to 1 g Avicel (0,14 mg/12,3 mg)
- Duration of incubation for CBD to bind to Avicel: about 30 minutes
- Washing and Lysis-buffer: 50mM Tris-HCl (pH 8.0)
- If needed: Elution with 80% ethylen-glycol (EG) or 1/5 Pellet to 4/5 EG (100%) of the overall volume.
- KRX culture of [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)-GFP_His] seems to have a green glow.
- Team Cultivation & Purification:
- Made competent P. pastoris GS115 cells.
- Fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012]
- Settings: fermenter: Bioengineering NFL22 (7 L), final volume: 6 L, LB-medium with 60 µg/mL chloramphenicol and 300 µg/mL ampicillin added, 37 °C, stirrer increased 2 % if the pO2 got below 30 %, airflow: 5 NL/m. Problem: stirrer cascade did not work at the beginning.
- Harvest and centrifugation of cultivation & fermentations 09/14. Store pellet at 4 °C.
- Team Activity Test:
- Today we got the chance to work with Team Immobilization again. The samples they gave us were supernatants from different immobilization approaches. The team tested diverse bead concentrations with even higher amounts of beads then before. Check their labjournal for further information about the output of the measurements. Besides the supernatants we also received beads with immobilized laccases TEVL0 and [http://partsregistry.org/Part:BBa_K863005 ECOL]. So finally we got to use the filter plates that helped us to carry out the activity measurements and then stop the reaction at a precice point of time via centrifugation. Team Immobilization gave us enough samples for 7 measurements after different reactions times. We chose to stop the reaction after 2, 4, 8, 16, 32 and 64 minutes, respectively, to check the behavior of the immobilized laccase in relation to the time. Check their lab journal for the very exciting results!
Sunday September 16th
- Team Cellulose Binding Domain:
- Colony-PCR of 15 colonies from the [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His] transformation-plate (biotaq - Armin Nestat's recipe) with the result of a lot of positive clones.
- Prepared the sequencing of three [http://partsregistry.org/Part:BBa_K863122 const.GFP_His], three [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] and [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg]
- Lysis of [http://partsregistry.org/Part:BBa_K863113 CBDcex(T7)+GFP_His] isn't glowing anymore, or never was.
- Team Cultivation & Purification:
- Harvest and centrifugation of fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012]. Store pellet at 4 °C.
- Cell disruption via sonification and purification of cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863103 BBa_K863103] via Ni-NTA column.
| 55px | | | | | | | | | | |
 "
"





