Team:MIT/Results
From 2012.igem.org
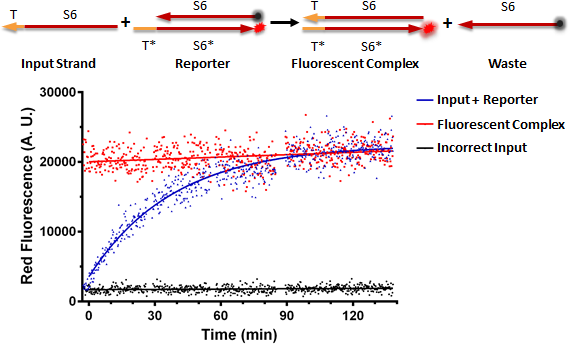
RNA Strand Displacement In Vitro

Nucleic Acid Delivery
Delivery of Plasmid DNA to Mammalian Cells
Delivery of 2'-O-Me RNA to Mammalian Cells
Delivery of Plasmid DNA which transcribes short RNA Inputs
Delivery of Plasmid DNA to Mammalian Cells
Delivery of 2'-O-Me RNA to Mammalian Cells
Delivery of Plasmid DNA which transcribes short RNA Inputs
In Vivo RNA Strand Displacement
Strategy 1: Lipofectamine 2000 Transfection of RNA version of Reporter from Winfree/QIan 2011 Paper
Images will go here from April 24th experiment - display red fluorescence in all wells, including those that only got reporter or the wrong input - also see red vesicles indicating reporter comes apart inside the vesicles
Strategy 2: Switch Transfection reagent to RNAiMAX
RNAiMAX is supposed to be a better transfection reagent for double stranded RNA
Images will go here from experiment from June 13th onward where we do not see red vesicles, however we still see whole cell red fluorescence
Strategy 3: Tag RNA strand with an Alexa Fluorophore to act as a transfection marker
Strategy 4: Create DNA plasmids driving transcription of RNA inputs, while transfecting RNA Reporter
Strategy 4: Nucleofect RNA reporter, RNA inputs
[Strategy 5]: Redesign RNA Reporter
Strategy 1: Lipofectamine 2000 Transfection of RNA version of Reporter from Winfree/QIan 2011 Paper
Images will go here from April 24th experiment - display red fluorescence in all wells, including those that only got reporter or the wrong input - also see red vesicles indicating reporter comes apart inside the vesicles
Strategy 2: Switch Transfection reagent to RNAiMAX
RNAiMAX is supposed to be a better transfection reagent for double stranded RNA
Images will go here from experiment from June 13th onward where we do not see red vesicles, however we still see whole cell red fluorescence
Strategy 3: Tag RNA strand with an Alexa Fluorophore to act as a transfection marker
Strategy 4: Create DNA plasmids driving transcription of RNA inputs, while transfecting RNA Reporter
Strategy 4: Nucleofect RNA reporter, RNA inputs
[Strategy 5]: Redesign RNA Reporter
 "
"

