Team:TU Munich/Notebook/Labjournal
From 2012.igem.org



June
Wednesday, June 13th
Exchange of the Multiple Cloning Site of pYES2
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pYES2
Hybridisation of the primers O1 with O2 and O3 with O4
Primer preparation:
- centrifugation
- dilution in the denoted quantity in bidest. water (concentration = 100 pmol/µl)
- centrifugation
Hybridisation:
| volume | reagent |
| 34 µl | ddH2O |
| 5 µl | PNK-buffer |
| 2.5 µl | Primer O1 |
| 2.5 µl | Primer O2 |
| 1 µl | PNK (10mM) |
| volume | reagent |
| 34 µl | ddH2O |
| 5 µl | PNK-buffer |
| 2.5 µl | Primer O3 |
| 2.5 µl | Primer O4 |
| 1 µl | PNK (10mM) |
- 30 min 37 °C
- 10 min 80 °C
- put the Thermo Block with the mixture in a styrofoam box and let it cool down over night
Thursday, June 14th
Exchange of the Multiple Cloning Site of pYES2
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pYES2
Digestion of pYES2 with HindIII and XbaI
| volume | reagent |
| 12 µl | Miniprep (pYES2 SH 1.7.3 with a concentration of 87.8 ng/µl) |
| 5 µl | NEB2 |
| 5 µl | 10x BSA |
| 2 µl | XbaI (10 U/µl) |
| 2 µl | HinIII (10U/µl) |
| 24 µl | ddH2O |
Incubation: 37 °C, 1.75 h
DNA preparative gel electrophoresis
- gel: 1% with LMP-agarose
- band 1: 10 µl DNA ladder (1kb)
- band 2: 50 µl probe + 5 µl loading dye
- 70 V, 90 min
Gelextration
- cut the bands (5.7-5.8 kb) and split it in two eppis
- m1=165.3 mg
- m2=211.1 mg
- QIAquick Gel Extractrion Kit
- eppi1: 495.9 µl QG-buffer
- eppi2: 633.3 µl QG-buffer
- step 6 was left out
- step 9: 30µl buffer, 4 min incubation
- the product was named P5
Transformation of plasmids from Prof. Schwab in E.coli XL-1 Blue
Investigator: Lara, Andrea
Aim of the experiment: Preparation of the plasmids for transformation
Overnight cultures of cells with limonenesynthase-plasmid from Prof. Schwab
- resuspend 50 µl / 200 µl of competent cells with 50 ml LB medium
- add 0,1 ml Ampicillin (100 µg/ml) and 0,28 ml Chloramphenicol (170 µ/ml) for strain 108
- add 0,07 ml Kanamycin (50 µg/ml) and 0,28 ml Chloramphenicol (170 µ/ml) for strain 106
- incubate at 37 °C
Friday, June 15th
Exchange of the Multiple Cloning Site of pYES2
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pYES2
Analytical DNA gel electrophoresis
- gel: 1.2 %
- band 1: 10 µl DNA ladder (1kb)
- band 2: 3 µl pYES2 digested + 7 µl TAE-buffer + 1 µl loading dye
- band 3: 3 µl O5 + 7 µl TAE-buffer + 1 µl loading dye
- band 4: 3 µl O6 + 7 µl TAE-buffer + 1 µl loading dye
- band 5: 10 µl DNA ladder (100 bp)
Ligation of Plasmid P5 (pYES2 digested) with the hybridized primers O5 and O6
| volume | reagent |
| 4 µl | pYES2 digested (P5) |
| 1 µl | O5 |
| 1 µl | O6 |
| 2 µl | T4-ligase buffer (10x) |
| 0,5 µl | T4 DNA-ligase |
| 11.5 µl | ddH2O |
Negative control
| volume | reagent |
| 4 µl | pYES2 digested (P5) |
| 2 µl | T4-ligase buffer (10x) |
| 0,5 µl | T4 DNA-ligase |
| 13.5 µl | ddH2O |
- water bath 16 °C
- after 3 h a probe for the transformation was taken
- the rest was ligated over the weekend
Transformation of E. coli with ligated products (P6)
- competent cells: SHXL1 Blue (by Simon)
- Transformation with ligation product (P6) and negative control
results:
- P6 (100 µl): 1 clone
- P6 (concentrated): 30 clones
- negative control (100 µl): 0 clones
- negative control (concentrated): 6 clones
Transformation of plasmids from Prof. Schwab into E.coli XL-1 Blue
Investigator: Andrea
Aim of the experiment: Preparation of the plasmids for transformation
Determination of the concentration with Nano Drop
| Sample | concentration [ng/µl] |
| P3 | 1353 |
| P4 | no result |
- the strain 106 culture was not grown satisfying and were incubated for 2 more days
Miniprep of pGex-4T-1 of strain 108 from Prof. Schwab
- see QIAprep Spin Miniprep Kit
- stored as P3 (-20 °C)
Sunday, June 17th
Exchange of the Multiple Cloning Site of pYES2
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pYES2
Picking clones for Miniprep
- 10 clones of transformed E.coli with P6 were picked
- medium: 5ml LB with Amp
Monday, June 18th
Exchange of the Multiple Cloning Site of pYES2
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pYES2
Miniprep of transformed E.coli with P6
- QIAprepS Spin Miniprep Kit
- step 3: invert 2-3 times (don't shake to avoid destruction of genomic DNA)
- the 10 Minipreps were named: P7 - P16
Determination of the concentration with Nano Drop
| Sample | concentration [ng/µl] | 260/280 |
| P7 | 157.6 | 2.32 |
| P8 | 207.3 | 1.63 |
| P9 | 171.2 | 2.02 |
| P10 | 183.1 | 1.57 |
| P11 | 160.4 | 2.2 |
| P12 | 179.9 | 1.75 |
| P13 | 179.2 | 2.07 |
| P14 | 188.3 | 1.6 |
| P15 | 166.7 | 2.05 |
| P16 | 174.6 | 2.08 |
Controll digestion with HindIII XbaI and NgoMIV
- Samples P7-P16: 2.5 µl
- Negative controll pYES SH 1.7.3: 2.5 µl
- Master Mix HindIII and XbaI: 17,5 µl for a 20 µl preparation
| volume | reagent |
| 3 µl | HindIII |
| 3 µl | XbaI |
| 24 µl | NEB2 |
| 24 µl | 10x BSA |
| 156 µl | ddH2O |
- Master Mix NgoMIV: 17,5 µl for a 20 µl preparation
| volume | reagent |
| 6 µl | NgoMIV |
| 24 µl | NEB4 |
| 180 µl | ddH2O |
Incubation: 37 °C, 1.5 h
Analytical gel electrophoresis of P7-P16
- gel: 1.5 %
- gel 1:
- band 1: 10 µl DNA ladder (1 kb)
- band 2: 3 µl pYES2 SH 1.7.3 digested with HindIII and XbaI + 7 µl TAE buffer + 1 µl loading dye
- band 3 - 12: 3 µl P7-P16 digested with HindIII and XbaI + 7 µl TAE buffer + 1 µl loading dye
- gel 2:
- band 1: 10 µl DNA ladder (1 kb)
- band 2: 3 µl pYES2 SH 1.7.3 digested with NgoMIV + 7 µl TAE buffer + 1 µl loading dye
- band 3 - 12: 3 µl P7-P16 digested with NgoMIV + 7 µl TAE buffer + 1 µl loading dye
Tuesday, June 19th
Transformation of BBa_I742111 (Limonenesynthase) into E.coli XL-1 Blue
Investigator: Andrea
Aim of the experiment: Transformation
- for each Biobrick 100 µl cells were used and pooled together with 2 µl of plasmid DNA
- Incubation on ice for 30 min
- 5 min heat shock at 37 °C
- cells were prefilled with 1 ml of LB-medium and incubated in a cell-culture shaker at 37 °C for 45 min
- 100 µl of these cell suspension were plated on antibiotic selection plates (Ampicillin)
- cell suspension was centrifuged at 13000 rpm for 60 s for resuspending the pellet with 100 µl LB and plating also
- incubation at 37 °C overnight
Wednesday, June 20th
Exchange of the Multiple Cloning Site of pYES2
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pYES2
Sequencing of P13 and P14: pYES2 with new MCS
- sequencing primer:
- 1.6 µM forward primer O9
- DNA P13 and P14
Transformation of BBa_I742111 (Limonenesynthase) into E.coli XL-1 Blue
Investigator: Daniela
Aim of the experiment: Transformation
Picking of Clones
- 6 clones were picked
- Incubation at 37 °C in LB + Amp
Thursday, June 21st
Transformation of BBa_I742111 and plasmids from Prof. Schwab into E.coli XL-1 Blue
Investigator: Lara, Andrea
Aim of the experiment: Controll of Transformation
Controll digestion
- Sample P3
| volume | reagent |
| 14 µl | Plasmid-DNA |
| 0,25 µl | NcoI |
| 2 µl | Buffer Tango (Fermentas) |
| 0,25 µl | HindIII |
| 2 µl | Buffer Red (Fermentas) |
| 1,5 µl | ddH2O |
- Sample P4
| volume | reagent |
| 6 µl | Plasmid-DNA |
| 0,25 µl | EcoRI |
| 2 µl | Buffer EcoRI (Fermentas) |
| 0,25 µl | NotI |
| 2 µl | Buffer Orange (Fermentas) |
| 9,5 µl | ddH2O |
- Sample Biobrick-clones
| volume | reagent |
| 5 µl | Plasmid-DNA |
| 0,25 µl | EcoRI |
| 2 µl | Buffer EcoRI (Fermentas) |
| 0,25 µl | PstI |
| 2 µl | Buffer Orange (Fermentas) |
| 10,5 µl | ddH2O |
Analytic Gelelectrophoresis
Friday, June 22nd
Transformation of E.coli XL1-Blue with pKS2µHyg-PAL-4Cl-CHS
Investigator: Ingmar, Volker
Aim of the experiment: Plasmid amplification
Operation Sequence:
- melting of 100 µl Ca-competent E.coli XL1-Blue cells
- addition of 1 µl of the Plasmid pKS2µHyg-PAL-4Cl-CHS
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Saturday, June 23rd
Quick Change mutagenis to remove NgoMIV from pYES2
Investigator: Ingmar, Volker
Aim of the experiment: Generation of an RFC 25 compatible version of the pYes2 Vector.
PCR
Reaction batch
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P7 pYes2_RFC25 MCS 1.1 template |
| 0.5 µl | 1:10 dilution of O38 (10 pmol/µL) |
| 0.5 µl | 1:10 dilution of O39 (10 pmol/µL) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 15 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 68°C | 6 min |
- The vector resulting from the PCR-product was named pYes2_RFC25 MCS 1.2.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells
- addition of 1 µl of the Plasmid P7 pYes2_RFC25 MCS 1.2
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Sunday, June 24th
Miniprep of E.coli XL1-Blue with pKS2µHyg-PAL-4Cl-CHS
Investigator: Ingmar, Volker
Aim of the experiment: Plasmid purification
Operation Sequence:
- A single clone of E.coli XL1-Blue with pKS2µHyg-PAL-4Cl-CHS was picked an transferred to 5 ml LB Amp on saturday evening. Incubation overnight at 37°C 180 rpm.
- Using a Quiagen kit a miniprep of the overnight culture was done.
Quick Change mutagenis to remove NgoMIV from pYES2
Investigator: Ingmar, Volker
Aim of the experiment: Removal of a NgoMIV restriction site in the backbone of pYes2.
Operational sequence:
- A single clone of E. coli pYes2_RFC25 MCS 1.2 was picked an transferred to 5 ml LB Amp. Incubation overnight at 37°C 180 rpm.
Transformation of 2 Biobricks into E. coli XL1-Blue
Investigator: Jeffery Truong
Aim of the experiment: Transformation of Biobricks into E. coli for plasmid propagation for PCR with new RFC pre- and suffix primer in order to do protein fusions.
- 2 Biobricks from the distribution kit were used: First, LexA (BBa_K105005, Plate 3 Well 9E) in the pSB1A2 plasmid with ampicillin resistance and second, the heme oxygenase (BBa_I15008, Plate 2 Well 13J) in the pSB2K3 plasmid with kanamycin resistance.
- 10 µL of autoclaved H2O were added to each well on the distribution kit. The well immediately turned red which means that one does it right.
- The now resuspended DNA liquids were transferred into a new ERG on ice.
- CaCL2 competent E. coli XL1-Blue cells from the stock were gently defrezed on ice.
- For each Biobrick 100 µL cells were used and pooled together with 2 µL of plasmid DNA in a ERG on ice.
- Incubation on ice for 30 min.
- 5 min heat shock at 37 °C.
- Each ERG now is transferred in a new ERG prefilled with 1 mL of LB-medium and incubated in a cell-culture shaker at 37 °C for 45 min.
- 100 µL of these cell suspension were plated on antibiotic selection plates (Ampicillin for LexA and Kanamycin for heme oxygenase).
- The rest of the cell suspension is centrifuged at 13000 rpm for 60 s and the supernatant is discarded.
- The pellet is resuspended with 100 µL for each ERG and is plated on another antibiotic selection plate
- These 4 plates were put at 37 °C overnight
Monday, June 25th
Miniprep of E. coli XL1-Blue with pYes2_RFC25 MCS 1.2
Investigator: Alois, Martin
Aim of the experiment: proof of successful removal of NgoMIV in the backbone of pYes2
Operation Sequence:
- Mini prep of pYes2 1.2. The resulting purified DNA is P33.
- Control digest of pYes2_RFC25 MCS 1.2 and p13 (+ analytical gel electrophoresis: 90 V, 1 h:
* 15 µl ddH20 * 2 µl NEBuffer4 * 0,5 µl NgoMIV * 2,5 µl pYes2 1.2/p13 * 37°C, 1 h.
Quick Change mutagenis to remove SpeI from pYES2_RFC25 MCS 1.2
Investigator: Ingmar, Volker
Aim of the experiment: Generation of an RFC 25 compatible version of the pYes2 Vector.
PCR
Reaction batch
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P33 template |
| 0.5 µl | 1:10 dilution of O44 (10 pmol/µL) |
| 0.5 µl | 1:10 dilution of O45 ((10 pmol/µL) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 15 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 68°C | 6 min |
- The procedure was furthermore applied to P13 and P14.
- The vector resulting from the PCR-product was named pYes2_RFC25 MCS 1.3.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- This operation sequence was applied to the PCR prducts of P33, P13 and P14 respectively.
- melting of 100 µl Ca-competent E.coli XL1-Blue cells
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Picking of E. coli cells on antibiotic selection plates: pSB1A2 plasmid with BBa_K105005 (LexA) and pSB2K3 plasmid BBa_I15008 (heme oxygenase)
Investigator: Jeffery Truong
Aim of the experiment: Picking colonies from transformed E. coli XL1-Blue, 4x picked for each Biobrick.
- pSB1A2 plasmid with BBa_K105005 (LexA): Colonies were on both ampicillin selection plates, the one with diluted cell suspension and the one with concentrated E. coli cell suspension. Typical E. coli colony morphology. Picking was performed on the plate with diluted cell suspension.
- pSB2K3 plasmid BBa_I15008 (heme oxygenase): Colonies were only on the kanamycin selection plate with concentrated cell suspension. The one with diluted susepension was empty. Typical but very small E. coli colonies. Picking was performed from the first plate.
- Picked pipette tips was transferred into a special cell-culture tubes with air-permeable, but sterile cover. In each tube 4 mL of LB-medium + ampicillin (???)(for pSB1A2) or kanamycin (35 mg/mL) (for pSB2K3).
- 4 colonies for each Biobrick was picked; total: 8 tubes overnight culture.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Tuesday, June 26th
Quick Change mutagenis to remove SpeI from pYes2_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Removal of a SpeI restriction site in the backbone of pYes2.
Operational sequence:
- For each transformation of the PCR-products of P14 and P33 a single clone was picked an transferred to 6 ml LB Amp. Incubation overday at 37°C 180 rpm. The transfomation with the PCR product of P13 was not successfull. Therfore no clone could be picked.
- Using a Quiagen kit a miniprep of the overnight culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
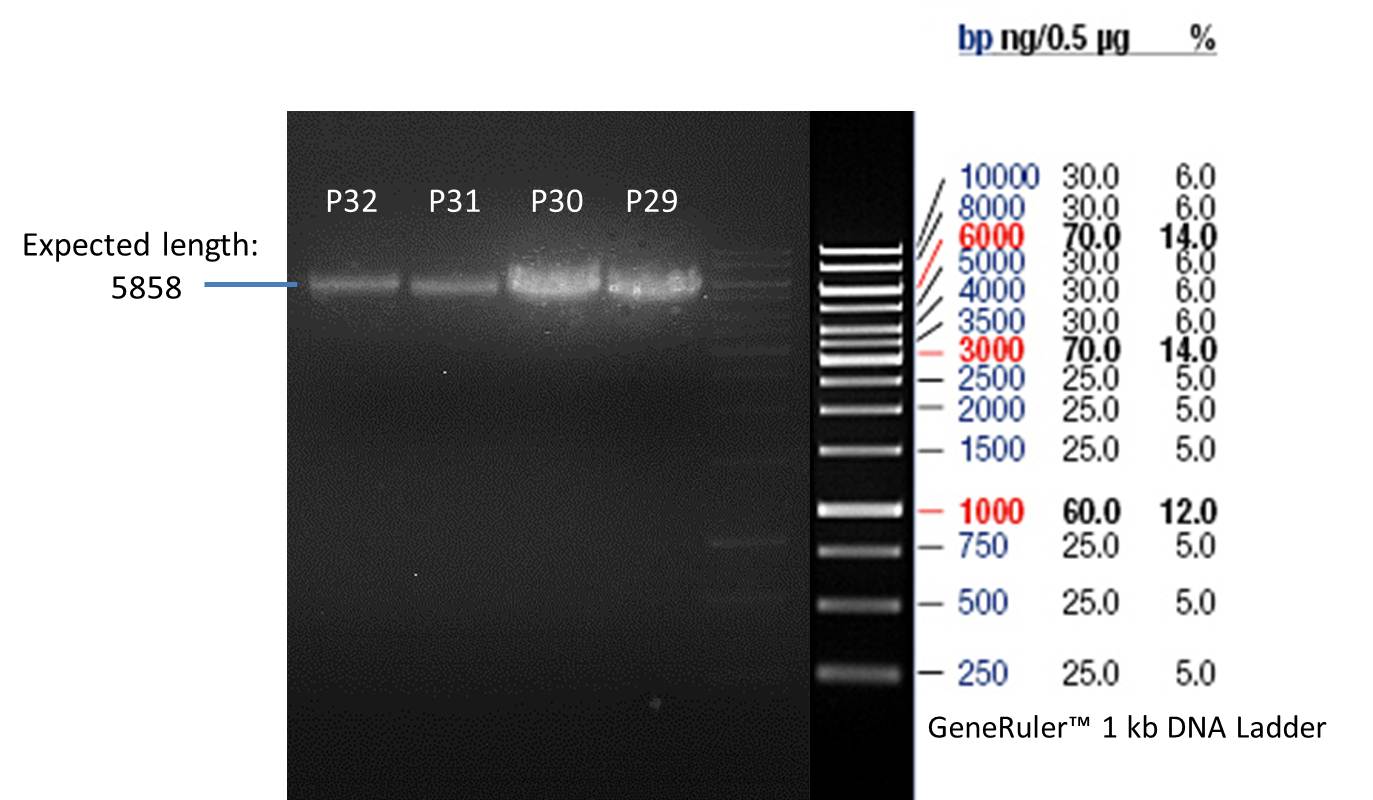
PCR product of P33(transformation done by Ingmar): P29
PCR product of P33(transformation done by Saskia&Jara): P30
PCR product of P14(transformation done by Ingmar): P31
PCR product of P14(transformation done by Saskia&Jara): P32
- Afterwards a control digestion of P29-P32 was done.
Reaction batch
| Plasmid | P29 | P30 | P31 | P32 |
| NEB4 buffer | 2 µl | 2 µl | 2 µl | 2 µl |
| DNA | 2,5 µl | 2,5 µl | 5 µl | 5 µl |
| SpeI-HF | 0.25 µl | 0.25 µl | 0.25 µl | 0.25 µl |
| NgoMIV | 0.25 µl | 0.25 µl | ||
| ddH2O | 15 µl | 15 µl | 12.75 µl | 12.75 µl |
| Sum | 20 µl | 20 µl | 20 µl | 20 µl |
- Incubation at 37 °C for 1h.
- Verification of control digest by agarose gel electrophoresis:
20 µl of each digest product was mixed with 2 µl DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V.
Verification of the PCR products P30, P31 and P33
Investigator: Saskia, Jara
Aim of the experiment: Verification of the PCR produts P30, P31 and P33
Nano Drop
| Sample | concentration [ng/µl] | 260/280 |
| P33 | 1072.6 | 1.01 |
| P30 | 1588 | 1.28 |
| P31 | 926.2 | 0.82 |
Analytical gel electrophoresis
- gel: 1 %
- band 1: 10 µl DNA ladder (1kb)
- band 2: P30
- band 3: P33
- band 4: P31
Control of the competent cells and transformation with P20
Investigator: Saskia, Jara
Aim of the experiment: Control of the competent cells and transformation with P20 Transformation
- competent cells: by Simon and Ingmar
- plasmid: P20
result:
- successful transformation: red colonies
PCR of PAL, 4CL, CHS, OMT (Coumaryl-CoA)
Investigator: Daniela, Mary
Determination of concentration of plasmids (Nanodrop): c(pKS2µHyg-PAL-4CL-CHS) = 500 ng/µl c (pOMT) = 20 ng/µl
PCR:
| Name of tube | Enzyme | consensus (+)/ consensless (-) | used Oligos |
| CHS - | CHS | - | O13 and O24 |
| CHS + | CHS | + | O23 and O24 |
| PAL - | PAL | - | O15 and O16 |
| PAL + | PAL | + | O22 and O16 |
| OMT - | OMT | - | O17 and O26 |
| OMT + | OMT | + | O25 and O26 |
| 4CL - | 4CL | - | O19 and O20 |
| 4CL + | 4CL | + | O21 and O20 |
Reaction batch
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 4 µl | dNTP's (each 2.5 mM) |
| 0.5 µl | Pfu Ultra II (2.5 U/µL) |
| 5 µl | 1:10 dilution of used forward primers (10µM) |
| 5 µl | 1:10 dilution of used reversed primers (10µM) |
| 1 µl | DNA (pKS2µHyg-PAL-4CL-CHS 50 ng/µL or pOMT 20 ng/µL) |
| 29.5 µL | bidest. sterile Water |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 5 min (and adding Pfu Ultra after 3 min) |
| 2 | 30 | 95°C | 30 sec |
| 46°C | 2.5 min | ||
| 72°C | 1.5 min | ||
| 3 | 72°C | 5 min |
PCR purification
- Purification was done using QIAquick PCR Purification Kit (250)
Analytical Gel Electrophoresis:
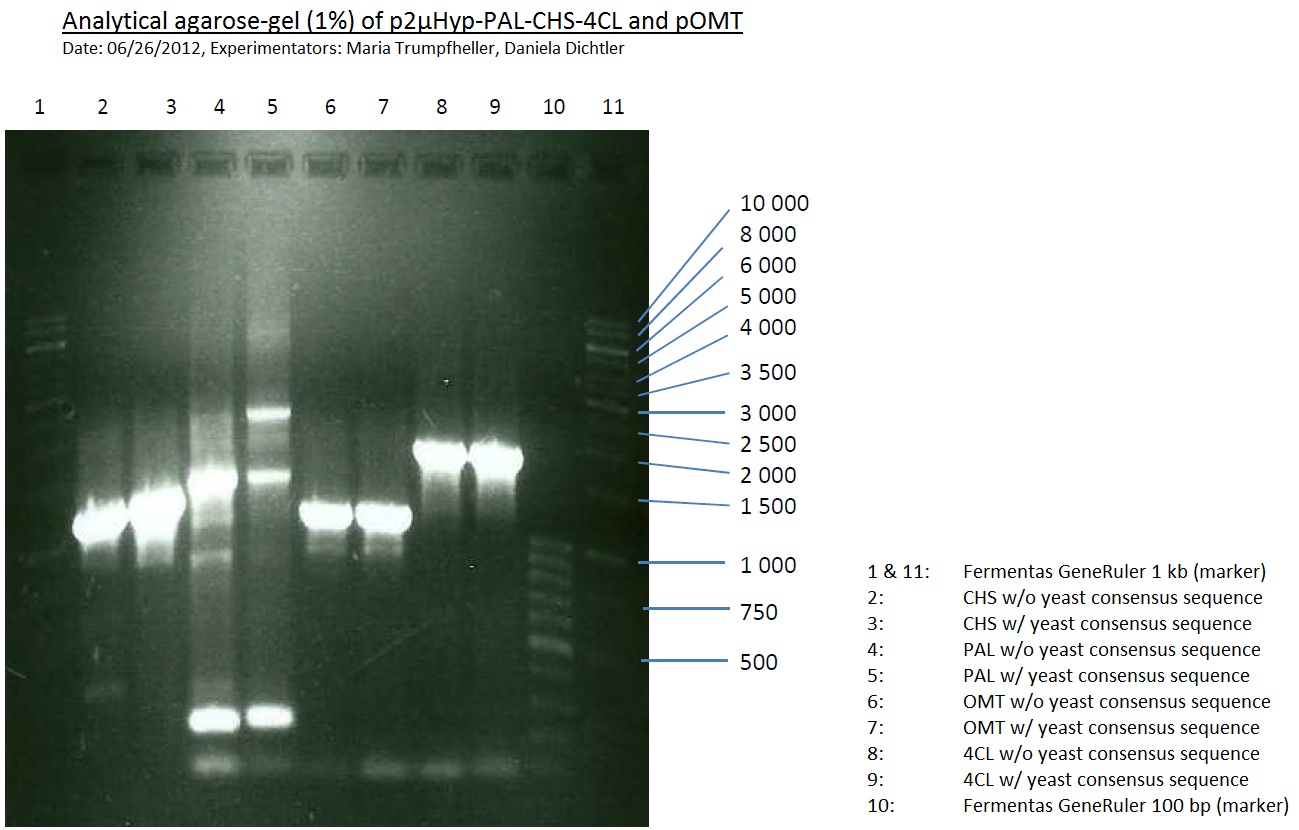
-> going on with CHS, 4CL and OMT; the PCR of PAL will be repeated
Miniprep and analytical gel of picked transformed overnight culture with pSB1A2 plasmid with BBa_K105005 (LexA) pSB2K3 plasmid BBa_I15008 (heme oxygenase)
Investigator: Jeffery Truong, Georg Schützinger
Aim of the experiment: Plasmid isolation from the picked transformed overnight E. coli cells with pSB1A2 plasmid with BBa_K105005 (LexA) pSB2K3 plasmid BBa_I15008 (heme oxygenase).
- The LB-medium with antibiotics of every tube was opaque which means that the picked cells were successfully inoculated.
- Centrifugation step at 5000 rpm for 10 min at 16 °C.
- Every single step now was performed on ice.
- Miniprep (Qiagen Qiaprep spin) after manufacturer's protocoll.
- Analytical restriction master mix was prepared after following scheme (Using XbaI and PstI):
- 4.4 µL XbaI
- 4.4 µL PstI
- 17.6 µL Tango-buffer (10x)
- 132 µL ELGA H2O
- 17.5 µL from the master mix was poooled together with 2.5 µL of plasmid DNA. That corresponds to 2.5 µL of plasmid DNA, 0.25 µL XbaI, 0.25 µL PstI, 2 µL Tango-buffer (10x), 15 µL ELGA H2O.
- Incubation at 37 °C for 120 min on a ERG heating unit.
- BUT: error was performed during preparing the digested plasmid DNA for analytical gelelectrophoresis in the dilution step. 1:10 dilution of analyctical probe with DNA loading buffer:
- 3.3 µL sample (contains already 1x loading buffer!)
- 0.7 µL loading buffer (?x)
- 6 µL TAE-buffer
- Should have done: 3 µL sample + 1 µL loading buffer (10x) + 6 µL TAE-buffer (1x)
- DNA-ladder preperation: 10 µL ladder stock solution + 10 µL DNA loading buffer + 80 µL TAE-buffer. 10 µL of this solution was pipetted in one gel pocket of the prepared 1% agarose gel including ehtiudiumbromid.
- 20 µL of each samples were also pipetted into the gel pockets.
- The gel pockets were pipetted after following scheme:
| Heme oxygenase (colony 1) | Heme oxygenase (colony 2) | Heme oxygenase (colony 3) | Heme oxygenase (colony 4) | DNA-ladder | LexA (colony 1) | LexA (colony 2) | LexA (colony 3) | LexA (colony 4) |
- Gel electrophoresis at 90 V
- After 20 min the resolution was still poor; 20 min longer.
- Analytical Gel okay, but samples interchanged
Friday, June 29th
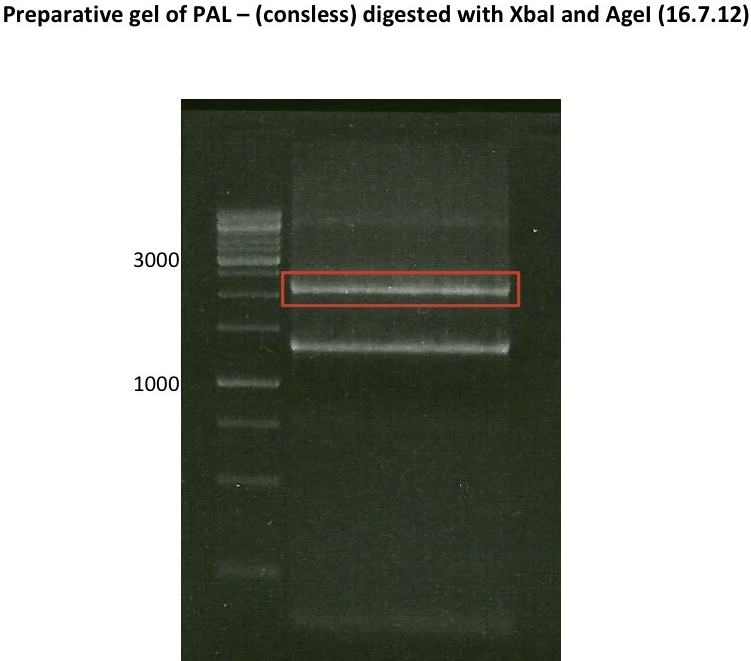
Preparative digest of PCR-products of 4CL, CHS and OMT
Investigator: Katrin, Mary
each digestion will dure 2.5h at 37°C
- CHS: digestion with Xba1 and HF-Age1 (both NEB)
| volume | reagent |
| 25µl | PCR-product |
| 5µl | Buffer NEB4 |
| 0.5µl | BSA |
| 1µl | Xba1 (NEB; 20u/µl) |
| 1µl | HF-Age1 (NEB; 20u/µl) |
| 17.5µl | bidest. sterile H2O |
- OMT: digestion with Xba1 and HF-Age1 (both NEB)
| volume | reagent |
| 25µl | PCR-product |
| 5µl | Buffer NEB4 |
| 0.5µl | BSA |
| 1µl | Xba1 (NEB; 20u/µl) |
| 1µl | HF-Age1 (NEB; 20u/µl) |
| 17.5µl | bidest. sterile H2O |
- 4CL: digestion with Xba1 and Pst1 (both Fermentas)
| volume | reagent |
| 25µl | PCR-product |
| 5µl | Buffer Tango |
| 2µl | Xba1 (Fermentas; 10u/µl) |
| 3µl | Pst1 (Fermentas; 10u/µl) |
| 15µl | bidest. sterile H2O |
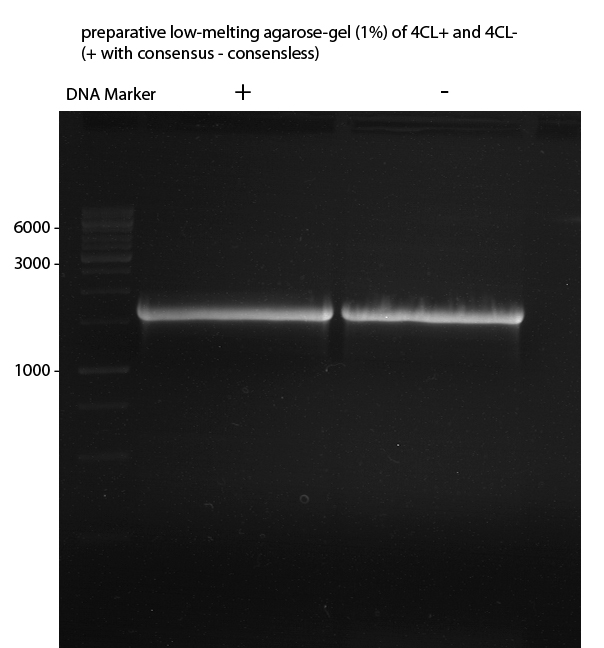
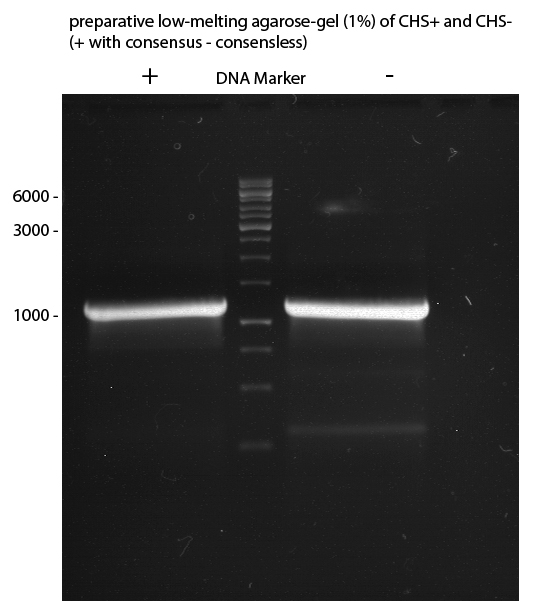
Preparative Gelelectrophoresis of PCR-products of 4CL, CHS
Investigator: Katrin, Mary
Gelextraction of 4CL+, 4CL-, CHS+, CHS- (bands are as expected; +=with consensus-sequence, -=without consensus-sequence)
DNA-purification with Kit from Quiagen
Quick Change mutagenesis to remove PstI in URA3 from pYES2_RFC25 MCS 1.2
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the pYes2 Vector.
PCR
Reaction batch
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P29 template |
| 0.5 µl | 1:10 dilution of O40 (10 pmol/µL) |
| 0.5 µl | 1:10 dilution of O41 ((10 pmol/µL) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Turbo DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 15 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6.5 min |
- The procedure was furthermore applied to P31.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- This operation sequence was applied to the PCR prducts of P29 and P31 respectively.
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Saturday, June 30th
Quick Change mutagenis to remove PstI in the URA 3 gene from pYes2_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Removal of a PstI restriction site in the backbone of pYes2.
Operational sequence:
- For each transformation of the PCR-products of P29 and P30 a single clone was picked an transferred to 6 ml LB Amp. Incubation overnight at 37°C 180 rpm. The transfomation with the PCR product of P31, P32 and P33 was not successfull. Therfore no clone could be picked from these plates and four instead of one clone was picked from the plates containing the transformations of P29.
July
Sunday, July 1st
Quick Change mutagenis to remove PstI in the URA 3 gene from pYes2_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Removal of a PstI restriction site in the backbone of pYes2.
Operational sequence:
- Using a Quiagen kit a miniprep of the overnight culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
1st PCR product of P29: P34
2nd PCR product of P29: P35
3rd PCR product of P29: P36
4th PCR product of P29: P37
PCR product of P30: P38
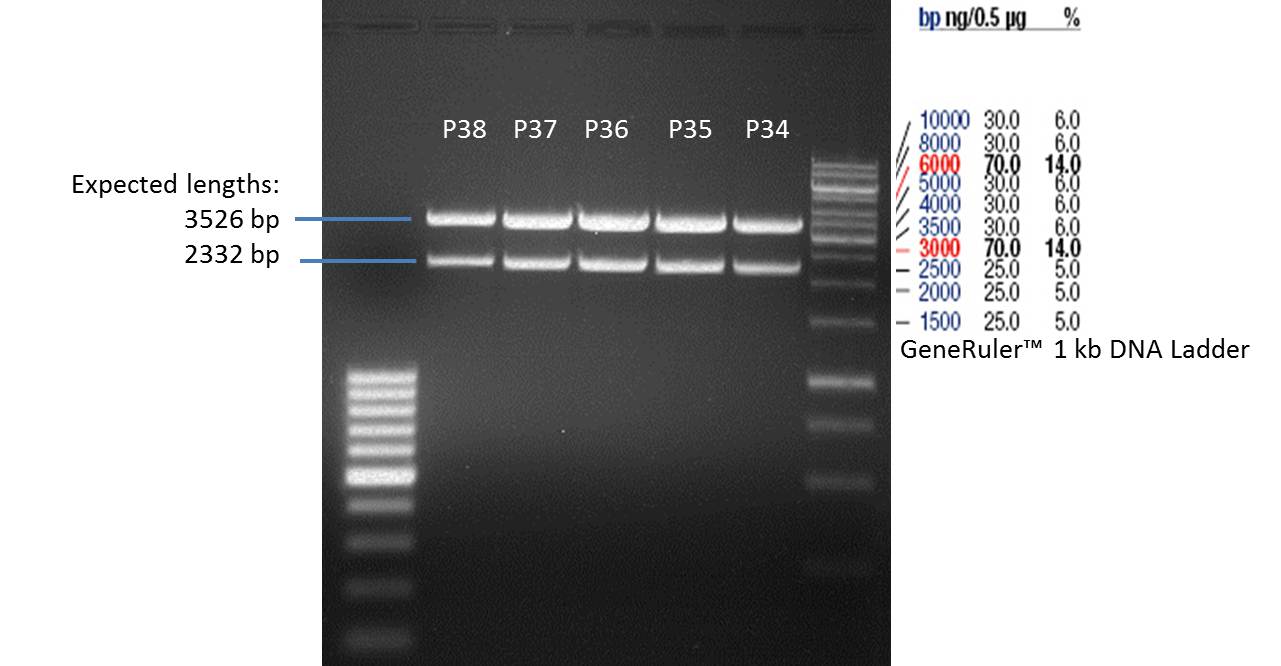
- Afterwards a control digestion of P34-P38 was done.
Reaction batch
| Plasmid | P34 | P35 | P36 | P37 | P38 |
| Fermentas 10x R buffer | 0.5 µl | 0.5 µl | 0.5 µl | 0.5 µl | 0.5 µl |
| DNA | 2 µl | 2 µl | 2 µl | 2 µl | 5 µl |
| PstI | 0.25 µl | 0.25 µl | 0.25 µl | 0.25 µl | 0.25 µl |
| ddH2O | 17.25 µl | 17.25 µl | 17.25 µl | 17.25 µl | 14.25 µl |
| Sum | 20 µl | 20 µl | 20 µl | 20 µl | 20 µl |
- Incubation at 37 °C for 1h.
- Verification of control digest by agarose gel electrophoresis:
20 µl of each digest product was mixed with 4 µl 6x DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V.
- All digest products show the expected two bonds at 3526 bp and at 2332 bp. The Miniprep product P35 was chosen to be used for the further Quickchange Mutagenesis.
Quick Change mutagenesis to remove PstI in the 2µ ori from pYES2_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the pYes2 Vector.
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P35 template |
| 0.5 µl | 1:10 dilution of O42 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P35 template |
| 0.5 µl | 1:10 dilution of O43 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Monday, July 2nd
Repetition of PCR of PAL
Investigator: Mary
Reaction batch
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 4 µl | dNTP's (each 2.5 mM) |
| 0.5 µl | Pfu Ultra II (2.5 U/µL) |
| 5 µl | 1:10 dilution of used forward primers (10µM) |
| 5 µl | 1:10 dilution of used reversed primers (10µM) |
| 1 µl | DNA (pKS2µHyg-PAL-4CL-CHS 50 ng/µL) |
| 29.5 µL | bidest. sterile Water |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 2 min (and adding Pfu Ultra after 2 min) |
| 2 | 30 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 72°C | 2.5 min | ||
| 3 | 72°C | 5 min |
PCR of LexA with primers including the RFC25 pre- and suffix
Investigator: Jeffery Truong, Georg Schützinger
Aim of the experiment: The Biobrick BBa_K105005 (LexA) has a RFC10 pre- and suffix, but we need RFC25 pre- and suffix for protein fusion, so we have to do a PCR with primer containing the RFC25 pre- and suffix.
- The received forward and reverse primer TUM12-LexA-fw and TUM12-LexA-rv are resuspended in 204 µL and 221 µL ELGA water as described in the data sheet to get a final primer concentration of 100 pmol/µL=100 µM. For the PCR reaction mixture we took 0.5 µL of these resuspended primer and add 4.5 µL of ELGA water to get a final primer concentration of 10 µM.
- Clone 3 of BBa_K105005 (LexA) has beed choosen for the PCR (ERG No. P23).
PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer |
| 1 µl | 10 µM Reverse Primer |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (BBa_K105005) from P23 (Clone 3) |
| 35.75 µL | ELGA Water |
| =50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 55 °C | 60 s | |
| 68 °C | 60 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | overnight |
Tuesday, July 3rd
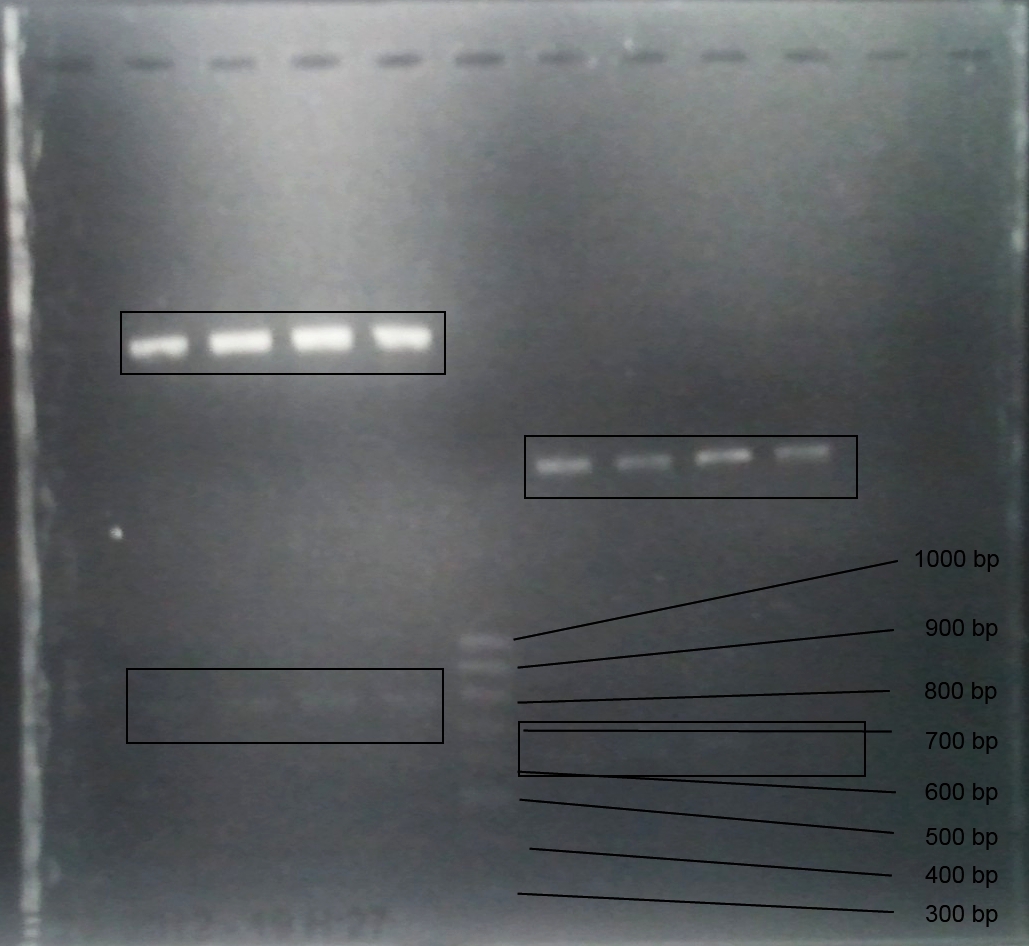
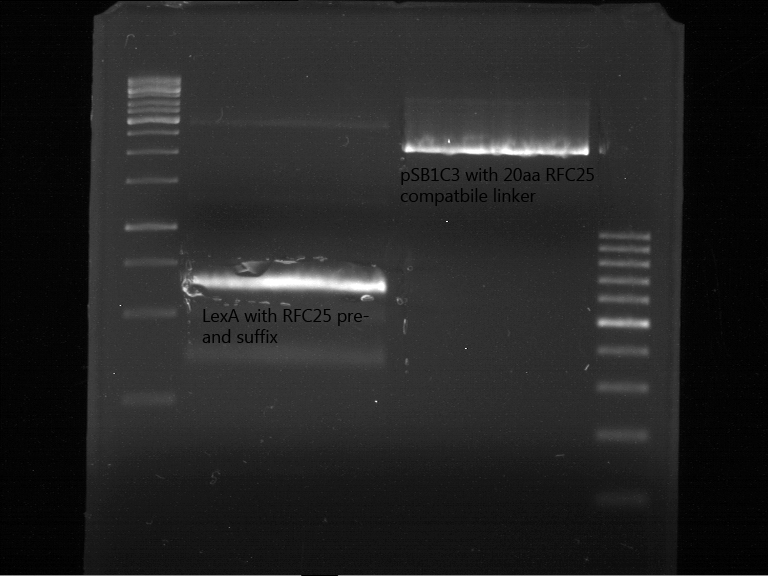
Analytic gelelectrophoresis of cleaned up PCR product from LexA with primer containing RFC25 pre- and suffix
Investigator: Jeffery Truong
Aim of the experiment: Analytical gelelectrophoresis of cleaned up PCR product from LexA (BBa_K105005) with primer containing the RFC25 pre- and suffix (TUM12-LexA-fw and TUM12-LexA-rv).
- The clean-up of the PCR product LexA (BBa_K105005) with primer containing the RFC25 pre- and suffix (TUM12-LexA-fw and TUM12-LexA-rv) was performed with the QIAquick PCR Purification Kit from Qiagen after manufacturer's protocoll.
- 5 µL of the purificated PCR product was taken to perform a analytical gelelectrophoresis to verify the success of the PCR.
- 1% agarose gel containing ethidium bromide was used for the analytical gelelectrophoresis.
- The analytical gelelectrophoresis was performed for 60 min at 90 V.
- Scheme of the gel:
| 100 bp ruler | PCR product of BBa_105005 | 1000 bp ruler |
LS: Plating of Schwab expression stains #106 & #108 which contain lavendula limonene synthase
Investigator: Lara Kuntz
Aim of the experiment: To get colonies of BL21 strains containing lavendula LS for amplification and subsequent plasmid extraction.
- Schwab strain #106 was plated on a chloramphenicol containing LB plate. #108 was plated on a amp+chlp containing LB plate. The plates were incubated at 37°C over night.
Quick Change mutagenis to remove PstI in the 2µ Ori from pYes2_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Removal of a PstI restriction site in the backbone of pYes2.
Operational sequence:
- From the transformation of the PCR-product of P35 two single clones were picked an transferred to 6 ml LB Amp. Incubation overnight at 37°C 180 rpm.
PCR of BBa_I742111 (Limonenesynthase) Clone 3 (Trafo 19.06.12)
Investigator: Andrea
PCR used forward primer with consensus sequence; PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O27 |
| 1 µl | 10 µM Reverse Primer O30 |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µl) |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 35.75 µl | dd water |
| 50 µL | TOTAL |
PCR used forward primer without consensus sequence; PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O28 |
| 1 µl | 10 µM Reverse Primer O30 |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µl) |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 35.75 µl | dd water |
| 50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 47 °C | 30 s | |
| 68 °C | 1,75 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | 1 h |
Analytical Gelelectrophoresis
- 5 µl DNA + 1 µl loading buffer
Wednesday, July 4th
Quick Change mutagenis to remove PstI in the 2µ Ori from pYes2_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Removal of a PstI restriction site in the backbone of pYes2.
Operational sequence:
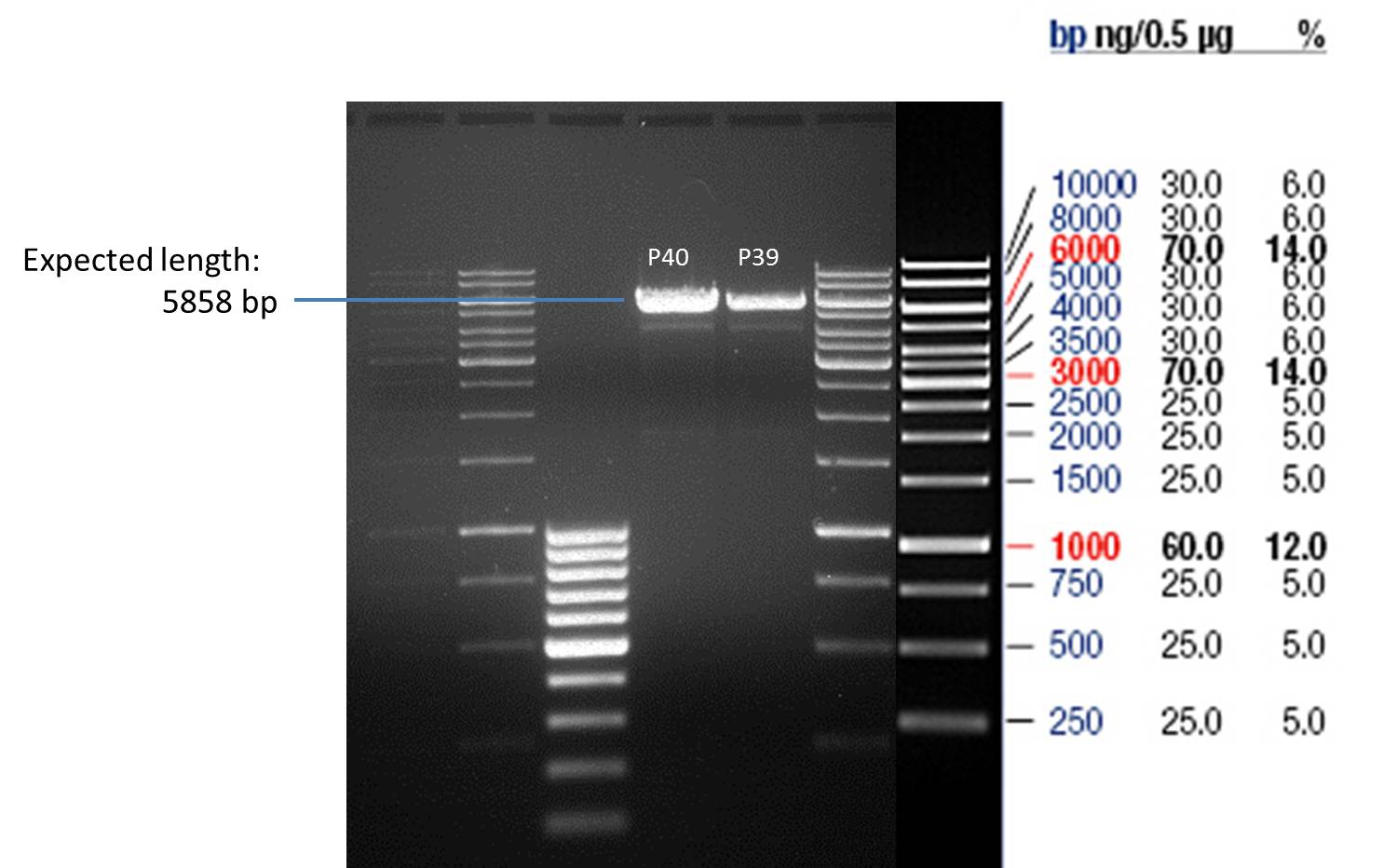
- Using a Quiagen kit a miniprep of the overnight culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
1st Transformation of P35: P43
2nd Transformation of P35: P44
- Afterwards a control digestion of P43 and P44 was done.
Reaction batch
| Plasmid | P43 | P44 |
| Fermentas 10x R buffer | 0.5 µl | 0.5 µl |
| DNA | 5 µl | 5 µl |
| PstI | 0.25 µl | 0.25 µl |
| ddH2O | 14.25 µl | 14.25 µl |
| Sum | 20 µl | 20 µl |
- Incubation at 37 °C for 1h.
- Verification of control digest by agarose gel electrophoresis:
20 µl of each digest product was mixed with 4 µl 6x DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V.
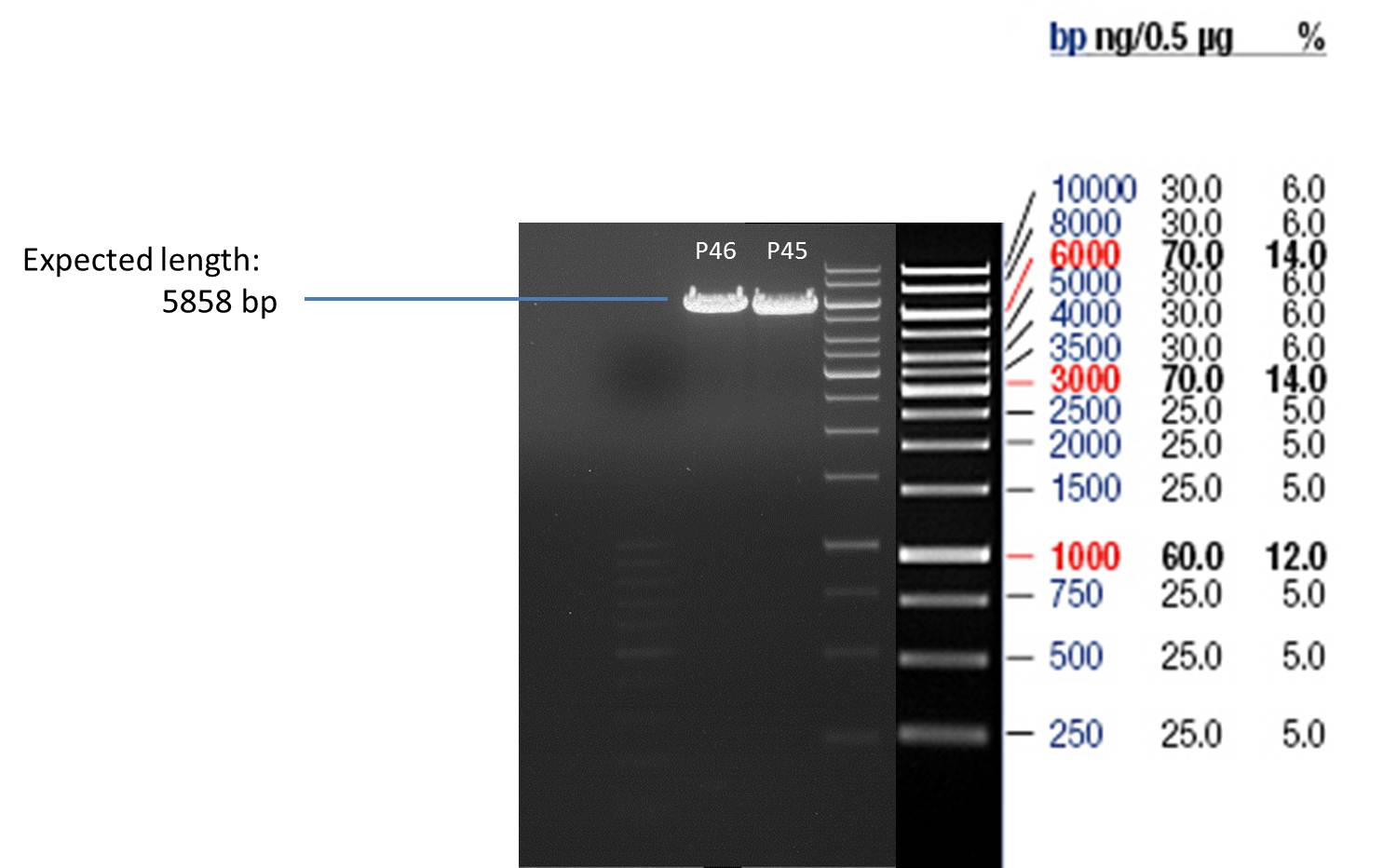
- All digest products show the expected bond at 5858 bp. The Miniprep product P40 was chosen to be used for the further Quickchange Mutagenesis.
Quick Change mutagenis to insert Ala in front of the Strep - tag II in pYES2_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the pYes2 Vector; operating purfication possibility via Strep-tag II.
PCR
Reaction batch
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P44 |
| 1 µl | 1:10 dilution of O54 (10 pmol/µL) |
| 1 µl | 1:10 dilution of O55 ((10 pmol/µL) |
| 16 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Turbo DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 16 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 68°C | 6 min |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Analytical Gelelektrophoresis of PCR-Products of PAL
Investigator: Mary
Aim of the experiment: purification and testing if PCR was successful
Purification of PCR-Products with purification kit from quiagen analytical gelelectrophoresis of PAL+, PAL-; expected band at 2,1 kb
Analytical Gel Electrophoresis: File:TUM12 20120704 PAL-PCR v2.tiff
-> PCR was not successful, no band at 2,1 kb
-> next steps: new Design of Primer and repetition of PCR with new primers
Picking of clones of Schwab expression stains #106 & #108
Investigator: Andrea
Aim of the experiment: Getting clones of cells with plasmids containing the gene for limonenesynthase and amplification of these clones for further plasmid preparation.
Picking of Clones
- 2 clones of every stain were picked
- Incubation at 37 °C in LB + Amp (#108) / LB + Kan (#106)
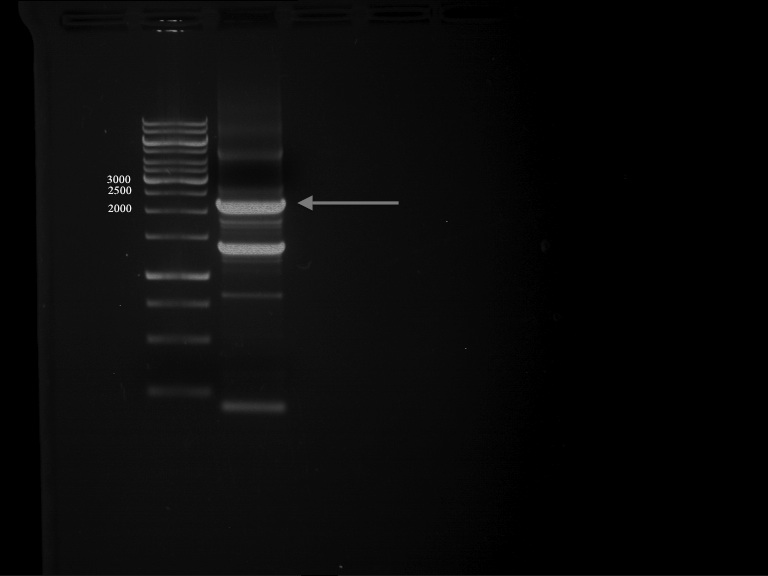
Preparative Gelelectrophoresis of PCR-products of OMT
Investigator: Mary, Katrin
Aim of the experiment: Purification of the previously digested DNA, test if digestion was successful
picture follows!
Gelextraction of OMT- and OMT+ (bands are as expected; +=with consensus-sequence, -=without consensus-sequence)
DNA-purification with Kit from Quiagen
Thursday, July 5th
Preparation of plasmids containing lavendula LS
Investigator: Lara
Aim: Purify Schwab plasmids containing lavendula limonene synthase
Experiment was conducted using Qiagen Plasmid Miniprep Kit.
P39: Plasmid from Schwab strain #106 (1); 35 ng/µl
P40: Plasmid from Schwab strain #106 (2); 72 ng/µl
P41: Plasmid from Schwab strain #108 (1); 240 ng/µl
P42: Plasmid from Schwab strain #108 (2); 120 ng/µl
Restriction digest of Schwab plasmids
Investigator: Lara
Aim: To check whether extracted plasmids from Schwab expression strains #106 & #108 are OK.
Digest of plasmid from strain #106 was conducted with the following protocoll:
500 ng Plasmid DNA
0,25 µl Nco1
0,25 µl Hind 3
2 µl Buffer Tango
dd H2O to a total volume of 20 µl.
Digest of plasmid from strain #108 was conducted with the following protocoll:
500 ng Plasmid DNA
0,25 µl EcoR1
0,25 µl Not1
2 µl Buffer Orange
dd H2O to a total volume of 20 µl.
(All enzymes and buffers were from Fermentas).
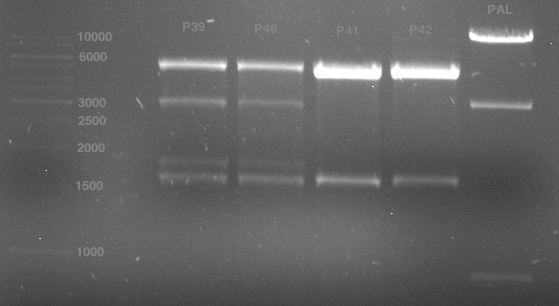
Analytical gel electrophoresis of digested Schwab plasmids
Investigator: Lara Aim: Check plasmids for insert.
Plasmids were digested, 2 µl loading dye (10x) was added to each sample. Gel was loaded in the following order:
1. 6 µl gene ruler 1 kb, 2.-6. 11 µl of: P39, P40, P41, P42, Coumaryl-Plasmid (Katrin)
Analytical Gelelektrophoresis of plasmid pKS2µHyg-PAL-4Cl-CHS
Investigator: Katrin
Aim of the experiment: testing if PAL is part of the plasmid that was sent to us (troubleshooting because PCR of PAL was not successful)
digestion took 1 h at 37°C
- digestion with ApaI (Fermentas)
| volume | reagent |
| 9.3 µl | plasmid pKS2µHyg-PAL-4Cl-CHS (miniprep) |
| 2 µl | Buffer B |
| 0.5 µl | ApaI (Fermentas; 10u/µl) |
| 8.2 µl | bidest. sterile H2O |
analytical gelelectrophoresis: expected band at 3,14 kb (Gal-PAL-XK)
-> experiment was successful, PAL is part of the plasmid pKS2µHyg-PAL-4Cl-CHS
Quick Change mutagenis to insert Ala in front of the Strep - tag II in pYES2_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the pYes2 Vector; operating purfication possibility via Strep-tag II.
Operational sequence:
- From the transformation of the PCR-product of P44 two single clones were picked an transferred to 6 ml LB Amp. Incubation overday at 37°C 180 rpm.
- Using a Quiagen kit a miniprep of the overnight culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
1st picked clone of P44: P45
2nd picked clone of P44: P46
Friday, July 6th
Quick Change mutagenis to insert Ala in front of the Strep - tag II in pYES2_RFC25 MCS
- Afterwards a control digestion of P45 and P46 was done.
Reaction batch
| Plasmid | P45 | P46 |
| Fermentas 10x O buffer | 2 µl | 2 µl |
| DNA | 3 µl | 3 µl |
| PstI | 0.25 µl | 0.25 µl |
| ddH2O | 14.75 µl | 14.75 µl |
| Sum | 20 µl | 20 µl |
- Incubation at 37 °C for 1h.
- Verification of control digest by agarose gel electrophoresis:
20 µl of each digest product was mixed with 4 µl 6x DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V.
- All digest products show the expected bonds at 5858 bp. The Miniprep product P45 was chosen to check the insertion of Ala in front of the strep-tag II via DNA sequencing.
The results of the sequencing are shown below:
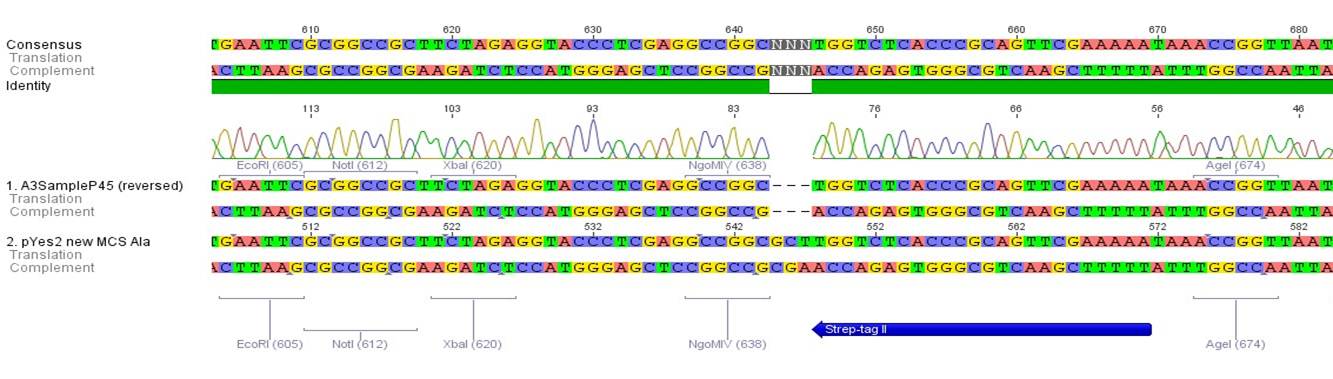 The sequencing results show that the insertion of Alanin in front of the Strep-tag II was not successful.
The sequencing results show that the insertion of Alanin in front of the Strep-tag II was not successful.
PCR of Schwab plasmid DNA to amplify gene for lavendula LS
Instructor: Lara
Aim: PCR of Schwab plasmids with primers which were designed to amplify the lavendula LS gene and to add RFC25 restriction sites.
3 different primer combinations were used:
1. O33/O37
2. O34/O37
3. O35/O37
Each primer combination was used for plasmid DNA amplification of P40 and P41.
PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer |
| 1 µl | 10 µM Reverse Primer |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Final: 1.25 units/50 µl) |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 35.75 µl | dd water |
| 50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 50 °C | 30 s | |
| 68 °C | 1,75 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | 1 h |
Saturday, July 7th
Quick Change mutagenesis to insert Ala in front of the Strep-tag II in pYES2_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the pYes2 Vector.
- As the sequencing of the first attempt to introduce Ala in front of the Strep-tag II did not show a successfull insertion of Ala, the quickchange mutagense was performed once again with a modified setup. Presumably a formation of primer dimers was responsable for the experiment's results. Therefore the second PCR was operated in two steps as shown below.
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P44 template |
| 0.5 µl | 1:10 dilution of O54 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Turbo DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P44 template |
| 0.5 µl | 1:10 dilution of O55 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Turbo DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Transformation of E.coli XL1 blue with ADH1 promoter, ADH1 terminator and TEF2 promoter
Investigator: Georg
Monday, July 9th
Phycocyanobilin (PCB) extraction from dried Spirulina platensis powder (part 1/4)
Investigator: Jeff, Alois, Martin
Aim of the experiment: Phycocyanobilin (PCB) is a cofactor neeeded for the funtion of phytochrome B. Phycocyanobilin is covalently bound to Cys457 of phytochrome B. Saccharomyces cerevisiae does not contain endogenous PCB. For proof of concept PCB should be added to the medium. In the following experiment, PCB is extracted from dried Spirulina platensis powder.
Operational sequence:
- 50 g of Spirulina platensis powder was (from concept-vitalprodukte.de) resuspended in 1.5 l of H2O (30 mg/l) in a beaker covered with aluminium foil.
- Stirring for 10 min at RT.
- Green Spirulina suspension was divided in 6 centrifuge bottles, covered in aluminium foil.
- Centrifation at 10500 rpm at 4 °C (SLA-3000 rotor, Thermo Scientific) for 1 h.
- Supernatant was discarded
- 25 ml of MeOH added to each bottle and was heavily shaked to resuspend the pellet for the next cleaning step with MeOH.
- Each bottle with the resuspended pellet were filled with MeOH to a final volume of 250 ml and shaked again to fully resuspend the pellet.
- Centrifugation at 10500 rpm at 4 °C (SLA-3000 rotor, Thermo Scientific) for 10 min.
- Supernatant was discarded.
- The last 4 steps were repeated until the supernatant of the washed pellet was colorless or cyanblue but not green anymore (Regulary, it takes 3 or 4 times). Pellet should be cyanblue now.
- The pellet of the 6 centrifuge tubes was collected in a sole centrifugation tube, covered in aluminium foil tube, by scratching it out with a small spoon.
- The remaining rest of the pellet which cannot be scratched out were resuspended in a small amount of MeOH and were transformed from tube to tube with a interim shaking step.
- This suspension was transferred into the tube with the scratched-out pellet.
- Centrifugation at 10500 rpm at 4 °C (SLA-3000 rotor, Thermo Scientific) for 10 min.
- Supernatant was discarded.
- Finally washed pellet was stored, wrapped in foil overnight for the methanolysis next day.
Tuesday, July 10th
Phycocyanobilin (PCB) extraction from dried Spirulina platensis powder (part 2/4)
Investigator: Jeff, Alois, Martin
Aim of the experiment: Phycocyanobilin (PCB) is a cofactor neeeded for the funtion of phytochrome B. Phycocyanobilin is covalently bound to Cys457 of phytochrome B. Saccharomyces cerevisiae does not contain endogenous PCB. For proof of concept PCB should be added to the medium. In the following experiment, PCB is extracted from dried Spirulina platensis powder.
Operational sequence:
- Washed pellet from the day before was resuspended in 500 ml MeOH.
- Heat suspension in a 500 ml flask in a water bath at 70 – 75 ºC with a condensing coil cooled with water for 5 – 8 hrs.
- After this, the suspension was transferred into a new centrifuge tube and centrifuged at 10500 rpm at 4 °C (SLA-3000 rotor, Thermo Scientific) for 20 min.
- The supernatant was decanted trough a filter paper into new centrifugation tube and stored, wrapped in a aluminium foil, at -20 °C.
- The pellets also was stored, wrapped in a aluminium foil, at -20 °C.
Plating of received E. coli containing biobricks
Investigator: Jeff
Aim of the experiment: The received biobricks were already transformed in E. coli and were in an agar stabs. These E. coli cells were transferred with an inoculation loop on antibiotic selection plates and were incubated over night.
Operational sequence:
- Bacterias containing plasmids with biobricks were transferred with a sterile inoculation loop on antibiotic plates and were incubated at 37 °C overnight.
- The biobricks were: BBa_K207001 in pSB1A2, BBa_K243006 in BBa_K157000, BBa_K300001 in K300007 (part for other subproject), BBa_K268000 in pSB6A0 (part for other subproject), BBa_K365005 RFC25 in pSB1C3, BBa_K365000 in pSB1C3, BBa_K207000 in pSB3K3, BBa_K165031 in pSB1AK3
Wednesday, July 11th
Phycocyanobilin (PCB) extraction from dried Spirulina platensis powder (part 3/4)
Investigator: Jeff, Alois, Martin
Aim of the experiment: Phycocyanobilin (PCB) is a cofactor neeeded for the funtion of phytochrome B. Phycocyanobilin is covalently bound to Cys457 of phytochrome B. Saccharomyces cerevisiae does not contain endogenous PCB. For proof of concept PCB should be added to the medium. In the following experiment, PCB is extracted from dried Spirulina platensis powder.
Operational sequence:
- The pellet after the first methanolysis of the day before was undergone a second methanolysis step to ensure high efficiacy of PCB extraction. Operational sequence was performed like the first methanolysis including the centrifugation and filtering step. The pellet after the second methanolysis should be more colorless and the filtered supernatant was freezed, like the one from the first methanolysis, at -20 °C, wrapped in aluminium foil
Transformation of E. coli XL1-Blue with pGADT7 and pGBKT7 plasmid for Yeast-two-hybrid (Y2H)
Investigator: Jeff
Aim of the experiment: pGADT7 and pGBKT7 are plasmids containing the transcriptional activation domain or the DNA binding domain of the transcription activator Gal4. pGADT7 contains the transcriptional activation domain of gal4; we want to clone the the first 100 residues of Pif3 (BBa_K365000) into this plasmid. As a result we have a fusion contruct of Gal4 and Pif3, which is nescassary for the light-switchable promoter system. pGBKT7 is for backup, if the ordered biobricks are not working.
Operational sequence:
- E. coli XL1-Blue are transformed with pGADT7 and pGBKT7 seperately after standard protocol of our lab.
Introducing new Saccharomyces cerevisiae strain (Y190 strain) from Schwab lab for Y2H
Investigator: Jeff
Aim of the experiment: The Y190 Saccharomyces cerevisiae is a special strain for Yeast-two-hybrid. This strain carries a Gal4 and Gal80 deletion to higher the signal/noise-ratio of protein-protein interactions. Reporter for protein-protein interactions are HIS3, lacZ and MEL1 and are encoded in the genomic DNA. Transformation markers are trp1, leu2 and cyhR2 and are encoded on the transformation plasmids.
Operational sequence:
- The freshly plated Y190 cells are transferred with a inoculation loop from the original plate on a new YPD agar plate.
- After 2 days the the plate was put in 4 °C.
Repetition of PCR of PAL
Investigator: Katrin, Daniela
Reaction batch
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 4 µl | dNTP's (each 2.5 mM) |
| 0.5 µl | Pfu Ultra II (2.5 U/µL) |
| 5 µl | 1:10 dilution of used forward primers (10µM) |
| 5 µl | 1:10 dilution of used reversed primers (10µM) |
| 1 µl | DNA (pKS2µHyg-PAL-4CL-CHS 50 ng/µL) |
| 29.5 µL | bidest. sterile Water |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 2 min (and adding Pfu Ultra after 2 min) |
| 2 | 30 | 95°C | 30 sec |
| 52°C | 1 min | ||
| 72°C | 2.5 min | ||
| 3 | 72°C | 5 min |
PCR purification
- Purification was done using QIAquick PCR Purification Kit (250)
Analytical gelelektrophoresis of PCR-Products of PAL
-> PCR was not successful, no band at 2,1 kb (picture follows)
Preparative Digest of pYES_iGEM
Investigator: Katrin, Daniela
Digestion of P50 with Xbal and NgOMIV
| volume | reagent |
| 10 µl | P50 (concentration: 264.5 ng/µl) |
| 2.5 µl | NEB4 |
| 2.5 µl | 10x BSA |
| 1 µl | XbaI (20 U/µl) |
| 2 µl | NgOMIV (10U/µl) |
| 7 µl | ddH2O |
Incubation: 37 °C, 3 h
Digestion of P50 with Xbal and PstI
| volume | reagent |
| 10 µl | P50 (concentration: 264.5 ng/µl) |
| 5 µl | Tango buffer |
| 2 µl | XbaI |
| 3 µl | PstI |
| 7.5 µl | ddH2O |
Incubation: 37 °C, 3 h
DNA preparative gel electrophoresis
- gel: 1% with LMP-agarose
- band 1: P50 digested with XbaI and PstI
- band 2: P50 digested with XbaI and NgOMIV
- 70 V, 90 min
Gelextration
- cut the bands
- QIAquick Gel Extractrion Kit was used
- step 6 was left out
- step 9: 30µl buffer, 4 min incubation
Ligation of digested P50 with digested PCR-products of PCR 15-20 (4CL, CHS and OMT)
Investigator: Katrin, Daniela
Concentration (Nano Drop:
4CL+ (PCR 15) = 40.3 ng/µl
4CL- (PCR 16) = 37.3 ng/µl
CHS+ (PCR 17) = 46.1 ng/µl
CHS- (PCR 18) = 51.2 ng/µl
OMT+ (PCR 19) = 22.3 ng/µl
OMT- (PCR 20) = 16.7 ng/µl
P50 digested with XbaI and PstI = 23.9 ng/µl (the digested Plasmid has the number P71)
P50 digested with XbaI and NgOMIV = 28.4 ng/µl (the digested Plasmid has the number P72)
- required volumes were calculated using Lab Tools
4CL+ (PCR 15) with P50 digested with XbaI and PstI
| volume | reagent |
| 5.27 µl | P50 digested |
| 2.73 µl | 4CL+ (PCR 15) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
4CL- (PCR 16) with P50 digested with XbaI and PstI
| volume | reagent |
| 5.13 µl | P50 digested |
| 2.87 µl | 4CL- (PCR 16) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
CHS+ (PCR 17) with P50 digested with XbaI and NgOMIV
| volume | reagent |
| 5.84 µl | P50 digested |
| 2.16 µl | CHS+ (PCR 17) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
CHS- (PCR 18) with P50 digested with XbaI and NgOMIV
| volume | reagent |
| 6 µl | P50 digested |
| 2 µl | CHS- (PCR 18) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT+ (PCR 19) with P50 digested with XbaI and NgOMIV
| volume | reagent |
| 4.45 µl | P50 digested |
| 3.55 µl | OMT+ (PCR 19) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT- (PCR 20) with P50 digested with XbaI and NgOMIV
| volume | reagent |
| 3.87 µl | P50 digested |
| 4,13 µl | OMT- (PCR 20) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
Negative control
| volume | reagent |
| 5 µl | P50 digested with XbaI and PstI or NgOMIV |
| 3 µl | ddH2O |
| 1 µl | T4 DNA-ligase |
| 1 µl | T4-ligase buffer (10x) |
- water bath 16 °C
Preparation of YPD medium
Investigator: A lot of Alois, Martin
Manual for YPD production
Yeast Extract Peptone Dextrose Medium (1 liter)
- 1% yeast extract
- 2% peptone
- 2% dextrose (D-glucose)
1. Dissolve the following in 1000 ml of water:
- 10 g yeast extract
- 20 g peptone
- 20 g dextrose (see note below if making plates)
2. Autoclave for 20 minutes on liquid cycle.
3. Store medium at room temperature. The shelf life is approximately one to two months.
Thursday, July 12th
Phycocyanobilin (PCB) extraction from dried Spirulina platensis powder (part 4/4)
Investigator: Martin, Jeff
Aim of the experiment: Phycocyanobilin (PCB) is a cofactor neeeded for the funtion of phytochrome B. Phycocyanobilin is covalently bound to Cys457 of phytochrome B. Saccharomyces cerevisiae does not contain endogenous PCB. For proof of concept PCB should be added to the medium. In the following experiment, PCB is extracted from dried Spirulina platensis powder.
Operational sequence:
- The supernatants (~1l in total) of the two previous experiments were pooled and put into a rotary evaporater in order to acquire a concentrate of approximately 100ml.
- The settings of the rotary evaporater: ~160 millibars, waterbath temperature of 25-30°C
- Furthermore, there were measures to be taken to protect the solution from direct sunlight: blinds down, aluminum foil wrapped around the waterbath
- Afterwards we transfered the Methanol solution into a separating funnel and added another 100ml of aqua dest..
- By means of adding chloroform we created two phases. The lower (chloroform) phase with the solved Phycocyanobilin was separated and put into another rotary evaporater flask. This step was repeated three times in order to get all the Phycocanobilin into the next step
- Then the chloroform was completely removed in the rotary evaporater (same settings as before), the pure (?) Phycocyanobilin in the flask was solved in 60 ml DMSO, transfered into a new flask and frozen at -20 °C.
Picking of transformated (pGADT7 and pGBKT7) E. coli cells on antibiotic selection plates
Investigator: Jeff
Aim of the experiment: pGADT7 and pGBKT7 are plasmids containing the transcriptional activation domain or the DNA binding domain of the transcription activator Gal4. pGADT7 contains the transcriptional activation domain of gal4; we want to clone the the first 100 residues of Pif3 (BBa_K365000) into this plasmid. As a result we have a fusion contruct of Gal4 and Pif3, which is nescassary for the light-switchable promoter system. pGBKT7 is for backup, if the ordered biobricks are not working.
Operational sequence:
- A E. coli colony was picked for each plasmid and was transferred in a tube containing 4 ml of LB-medium containing antibiotics (Amp for pGADT7 and Kan for pGBKT7).
- Overnight culture at 37 °C in a cell-culture shaker.
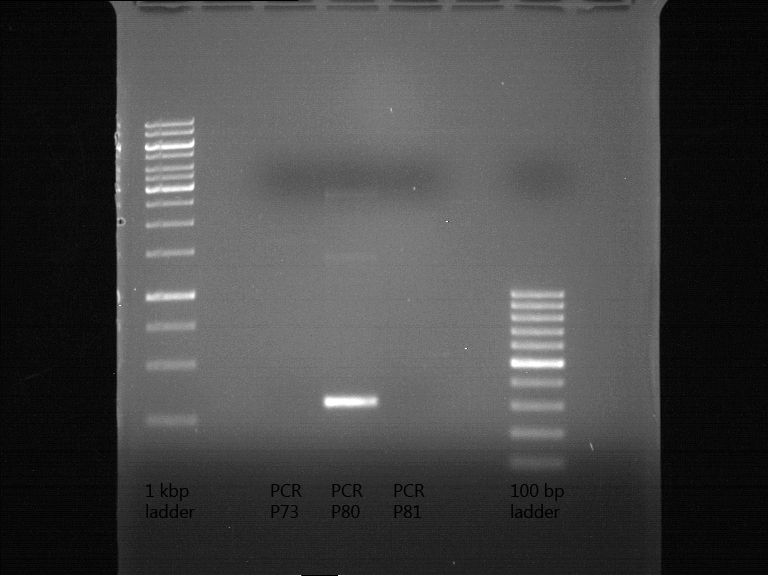
Miniprep of transformated E. coli from overnight culture (8 plasmids containing biobricks)
Investigator: Andrea
Aim of the experiment: Miniprep of transformated E. coli from overnight culture to get the plasmids with biobricks
NanoDrop Measure
| Plasmid | Concentration |
| P73 | 41.6 ng/µl |
| P74 | 85.2 ng/µl |
| P75 | 72.4 ng/µl |
| P76 | 254.3 ng/µl |
| P77 | 74.4 ng/µl |
| P78 | 50.0 ng/µl |
| P79 | 46.2 ng/µl |
| P80 | 116.7 ng/µl |
| P81 | 77.1 ng/µl |
| P82 | 65.5 ng/µl |
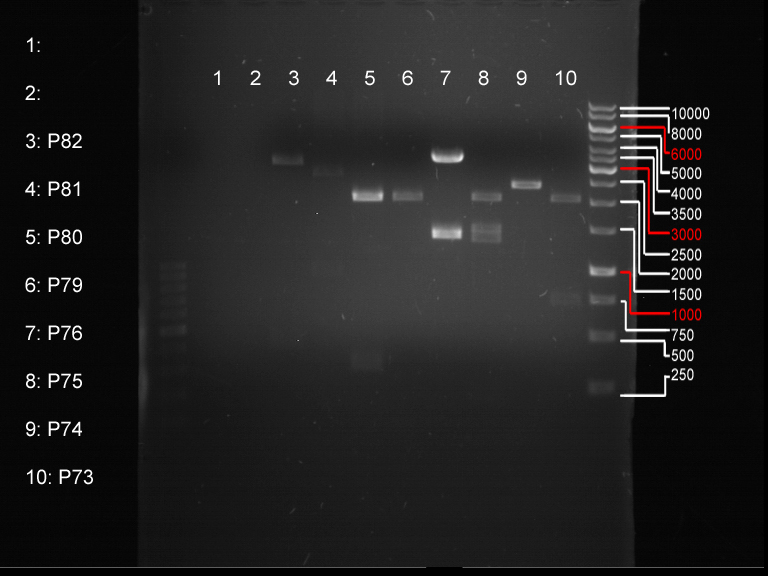
Analytical digestion and gelelectrophoresis of biobricks (8 plasmids containing biobricks)
Investigator: Jeff
Aim of the experiment: Checking whether the biobricks are part of the plasmid-backbone.
Operational sequence:
- Reaction batch for each plasmid:
| Reagent | Volume in µl |
| Tango Buffer 10x | 4 µl |
| XbaI (Fermentas) | 0.25 µl |
| PstI (Fermentas) | 0.5 µl |
| Plasmid DNA | 2.5 µl |
| ddH2O | 12.75 µl |
| TOTAL | 20 µl |
- Incubation at 37 °C for 1 h 45 min.
- Analytical gelelectrophoresis at 90 V for 1 h.
- Order of gel-pockets:
| 1 kbp ladder | P73 | P74 | P75 | P76 | P79 | P80 | P81 | P82 | 100 bp ladder |
| BAD | OK | OK | OK | OK | OK | BAD | OK |
- P73 and P81, both parts from Havard university are bad. To exclude errors, one should pick another colony for each plasmid and do again the analytical digestion and gelelectrophoresis after the miniprep.
Transformation of Ligationproducts of pYES2 + OMT, 4Cl and CHS in E.coli
Investigator: Mary
- adding 5µl ligation product in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (Withouth antibiotica) and incubate at 37°C, 30 min, 180 rpm
- plate on agar with Ampicillin over night
Repetition of PCR of PAL
Investigator: Daniela
Aim of the experiment: Repetition of PCR of PAL (so far not successful) with the use of different polymerase and try of 3 temperatures
only with PAL+, 3 different temperatures (see cycling parameters) and one batch at 52.5 °C with DMSO
Reaction batch
| volume | reagent |
| 10 µl | 5x Herculase II reaction buffer |
| 0,5 µl | dNTP Mix (each dNTP 2.5 mM) |
| 1 µl | Herculase II fusion DNA Polymerase |
| 1,25 µl | 1:10 dilution of used forward primers (O22) |
| 1,25 µl | 1:10 dilution of used reversed primers (O59) |
| 1 µl | 1:10 dilution of DNA (P19=pKS2µHyg-PAL-4CL-CHS 50 ng/µL) -> 5 ng/µL |
| 35 µL | ddH2O |
Reaction batch with DMSO
| volume | reagent |
| 10 µl | 5x Herculase II reaction buffer |
| 0,5 µl | dNTP Mix (each dNTP 2.5 mM) |
| 1 µl | Herculase II fusion DNA Polymerase |
| 1,25 µl | 1:10 dilution of used forward primers (O22) |
| 1,25 µl | 1:10 dilution of used reversed primers (O59) |
| 1 µl | 1:10 dilution of DNA (pKS2µHyg-PAL-4CL-CHS 50 ng/µL) -> 5 ng/µL |
| 1,5 µl | DMSO (3% des Ansatzes) |
| 33.5 µL | ddH2O |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 2 min |
| 2 | 30 | 95°C | 30 sec |
| 52.5°C | 30 sec | ||
| 72°C | 4 min | ||
| 3 | 72°C | 3 min |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 2 min |
| 2 | 30 | 95°C | 30 sec |
| 45°C | 30 sec | ||
| 72°C | 4 min | ||
| 3 | 72°C | 3 min |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 2 min |
| 2 | 30 | 95°C | 30 sec |
| 56.2°C | 30 sec | ||
| 72°C | 4 min | ||
| 3 | 72°C | 3 min |
Analytical gel electrophoresis
- no bands at all (picture follows)
- possibly due to low concentration of P19 (5 ng/µl)
Ligation of P93 (pSB1C3 digested with XbaI and AgeI) with digested PCR-products of PCR 17-20 (CHS and OMT)
Investigator: Daniela
Concentration (Nano Drop:
CHS+ (PCR 17) = 46.1 ng/µl
CHS- (PCR 18) = 51.2 ng/µl
OMT+ (PCR 19) = 22.3 ng/µl
OMT- (PCR 20) = 16.7 ng/µl
P93 (digested with XbaI and AgeI) = 4.8 ng/µl
CHS+ (PCR17) with P93 digested with XbaI and AgeI
| volume | reagent |
| 7.04 µl | P93 |
| 0.96 µl | CHS+ (PCR 17) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
CHS- (PCR 18) with P93 digested with XbaI and AgeI
| volume | reagent |
| 7.13 µl | P93 |
| 0.87 µl | CHS- (PCR18) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT+ (PCR19) with P93 digested with XbaI and AgeI
| volume | reagent |
| 6.19 µl | P93 |
| 1.81 µl | OMT+ (PCR19) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT- (PCR20) with P50 digested with XbaI and AgeI
| volume | reagent |
| 5.75 µl | P93 |
| 2.25 µl | OMT- (PCR20) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
Negative control
| volume | reagent |
| 5 µl | P93 |
| 3 µl | ddH2O |
| 1 µl | T4 DNA-ligase |
| 1 µl | T4-ligase buffer (10x) |
- water bath 16 °C
- products were named P94-P97
Preparative digest of PCR1-PCR7 and P50
Investigator: Andrea
Digestion of P50 with XbaI and NgoMIV
| volume | reagent |
| 20 µl | P50 |
| 4 µl | NEB Buffer |
| 0.4 µl | BSA |
| 1 µl | XbaI (10 U/µl) |
| 2 µl | NgoMIV (10 U/µl) |
| 27.4 µl | ddH2O |
Digestion of PCR1-PCR7 with XbaI and AgeI
| volume | reagent |
| 25 µl | PCR-Product |
| 5 µl | NEB Buffer |
| 0.5 µl | BSA |
| 1 µl | XbaI (10 U/µl) |
| 1 µl | AgeI (10 U/µl) |
| 32.5 µl | ddH2O |
Incubation: 37 °C, 3 h
Ligation of digested PCR1-PCR7 (digested with XbaI and AgeI) with pSB1C3 (digested with XbaI and AgeI) and with pYESnew (digested with XbaI and NgoMIV)
Investigator: Andrea
Concentration (Nano Drop:
LIMS Citrus (PCR 1) = 13.4 ng/µl
LIMS Citrus (PCR 2) = 10.3 ng/µl
LIMS Lavendula (PCR 3) = 2.2 ng/µl
LIMS Lavendula (PCR 3) = 9.9 ng/µl
LIMS Lavendula (PCR 3) = 9.6 ng/µl
LIMS Lavendula (PCR 3) = 9.8 ng/µl
LIMS Lavendula (PCR 3) = 12.3 ng/µl
P50 (digested with XbaI and NgoMIV) = 10.3 ng/µl
P93 (digested with XbaI and AgeI) = 4 ng/µl
PCR1 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.89µl | P50 |
| 3.11 µl | PCR 1 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR2 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.38 µl | P50 |
| 3.62 µl | PCR2 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR3 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 1.64 µl | P50 |
| 6.36 µl | PCR3 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR4 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.3 µl | P50 |
| 3.7 µl | PCR4 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR5 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.26 µl | P50 |
| 3.74 µl | PCR5 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR6 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.26 µl | P50 |
| 3.74 µl | PCR6 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR7 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 7.83 µl | P50 |
| 33.27 µl | PCR7 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
|}Negative control
| volume | reagent |
| 5 µl | P50 |
| 3 µl | ddH2O |
| 1 µl | T4 DNA-ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR1 with P93 digested with XbaI and AgeI
| volume | reagent |
| 4.89 µl | P93 |
| 3.11 µl | PCR1 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR2 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.38 µl | P93 |
| 3.62 µl | PCR2 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR3 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 1.52 µl | P93 |
| 6.48 µl | PCR3 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR4 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.3 µl | P93 |
| 3.7 µl | PCR4 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR5 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.26 µl | P93 |
| 3.74 µl | PCR5 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR6 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.26 µl | P93 |
| 3.74 µl | PCR6 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR7 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.72 µl | P93 |
| 3.28 µl | PCR7 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
- water bath 16 °C
- products were named P100-P115
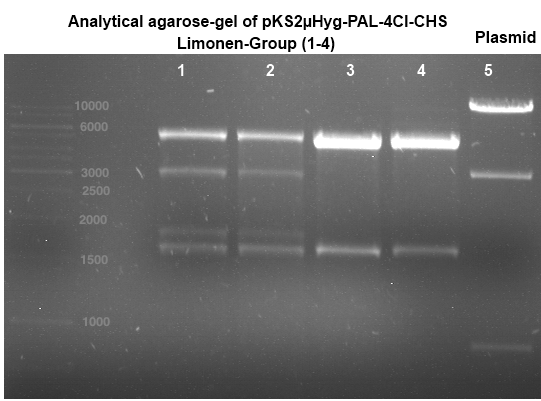
Analytical gel electrophoresis
Investigator: Andrea
Aim of the experiment: Analytical gel electrophoresis of products from restriction digest of plasmids PCR1 - PCR7 and PCR9 and P77, P78
File:C:\Users\Andrea\Dropbox\iGEM Teamordner\Gelfotos\Limonen\Bild1.png
Friday, July 13th
PCR of P73, P80 and P81 to add RFC25 pre- and suffix
Investigator: Jeff, Saskia
Miniprep, analcytical digestion and gelelectrophoresis of pGADT7 and pGBKT7
Investigator: Jeff, Saskia
Aim of the experiment:
Operational sequence:
Preparative digest of P19
Investigator:Daniela, Ingmar
Digestion of P19 with ApaI
| volume | reagent |
| 20 µl | P19 (concentration: 53.8 ng/µl) |
| 4 µl | Buffer B |
| 2 µl | ApaI (10 U/µl) |
| 14 µl | ddH2O |
Incubation: 37 °C, 3 h
DNA preparative gel electrophoresis
- gel: 1% with LMP-agarose
- band 1: P50 digested with XbaI and PstI
- band 2: P50 digested with XbaI and NgOMIV
- 70 V, 90 min
Gelextration
- cut the bands
- QIAquick Gel Extractrion Kit was used
- step 6 was left out
- step 9: 30µl buffer, 4 min incubation
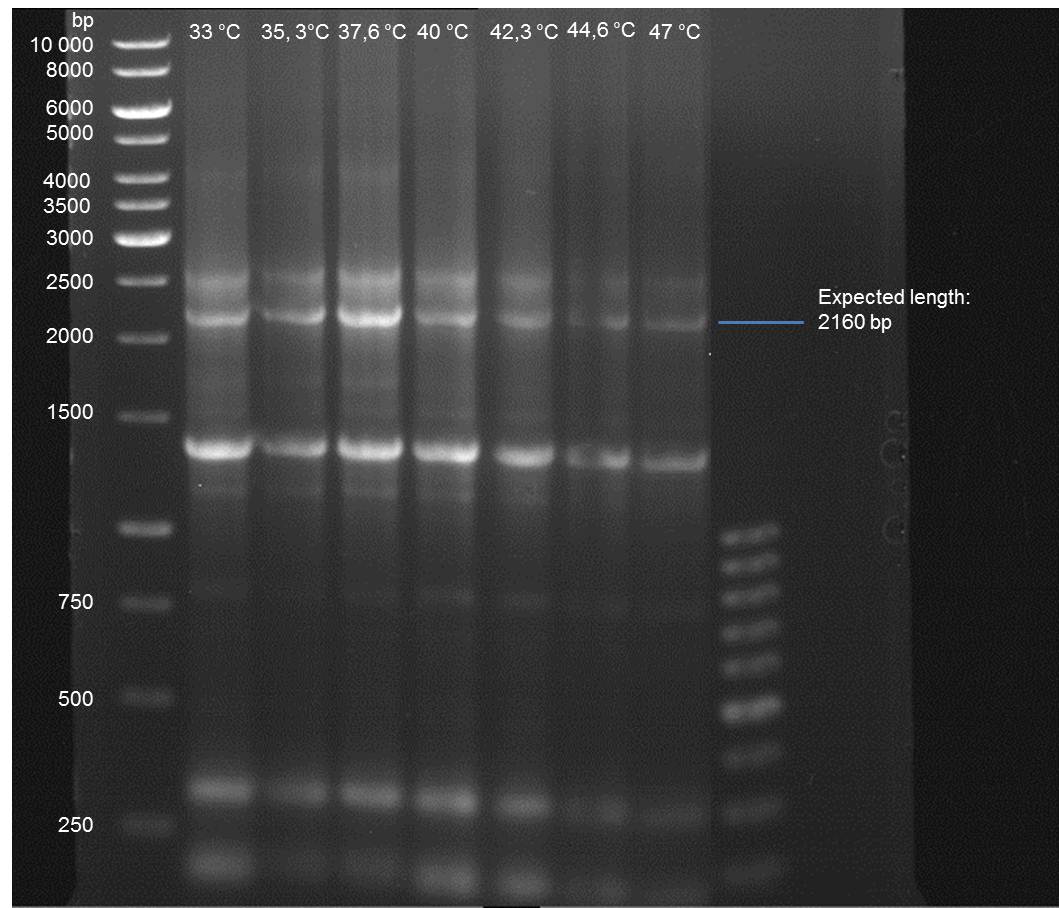
Gradient PCR of PAL to optimize the primer annealing temperature
Investigator: Ingmar
Aim of the experiment: As all PCR experiments to amplify the PAL gene contained in P19 failed, we decided to run a gradient PCR to optimize the primer annealing temperature.
PCR
Reaction batch
| volume | reagent |
| 1 µl | 10x Pfu Ultra II buffer |
| 0.5 µl | Plasmid P19 template |
| 0.5 µl | 1:10 dilution of O15 (10 pmol/µL) |
| 0.5 µl | 1:10 dilution of O59 (10 pmol/µL) |
| 0.25 µl | dNTP mix |
| 7 µl | ddH2O |
| 0.25 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 29 | 95°C | 30 sec |
| gadient 33-47.5 °C | 1 min | ||
| 68°C | 3 min | ||
| 3 | 1 | 68°C | 5 min |
| 4 | 1 | 4°C | infinity |
- Verification of the PCR by agarose gel electrophoresis:
10 µl of each PCR tube was mixed with 2 µl 6x DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V in a 1% agarose gel.
- A bond at the expected length of 2160 bp appears at 47 °C. Therefore a PCR using this primer annealing temperature will done on sunday.
Transformation of P94 - P97 (ligationproducts of pSB1C3 with CHS and OMT respectively) in E.coli
Investigator: Daniela
- adding 5µl ligation product (P94-P97) in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (Withouth antibiotica) and incubate at 37°C, 30 min, 180 rpm
- plate on agar with Ampicillin over night
wrong antibiotica was used!!! Repetition follows!!! Nevertheless, some colonies could be observed.
Ligation of LIMS in pYESnew and pSB1C3
Investigator:Andrea
Transformation
- adding 5µl ligation product (PCR1-PCR7) in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (without antibiotica) and incubate at 37°C, 30 min, 180 rpm
- plate on agar with Ampicillin (pYESnew) or Chloramphenicol (pSB1C3) over night
Sunday, July 15th
PCR purification of PCR products of P73, P80 and P81
Investigator: Jeff
Aim of the experiment: PCR was performed to introduce restriction sites to the gene of interest. MfeI and BamHI for P73 and RFC25 pre- and suffix for P80 and P81. The aim is to contruct fusion proteins with the aid of these restriction sites.
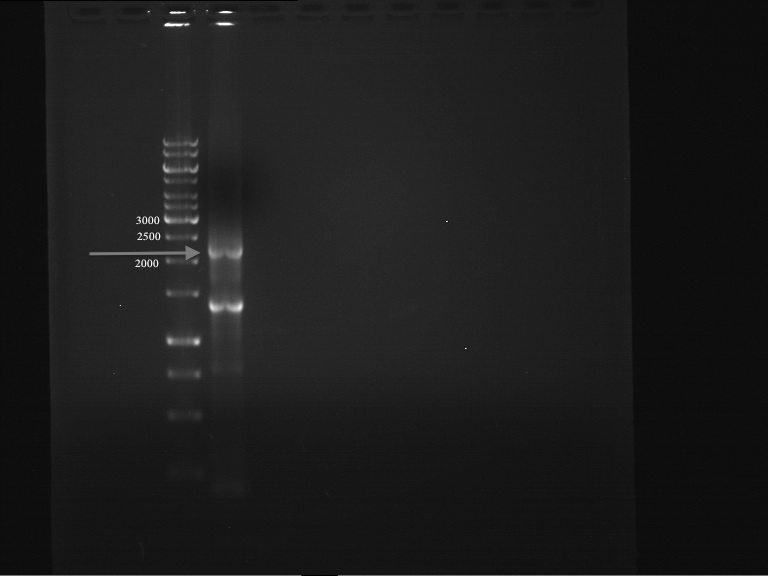
PCR of PAL consless using the optimized primer annealing temperature of 47 °C
Investigator: Ingmar
Aim of the experiment: Amplification of the PAL gene contained in P19.
PCR
Reaction batch
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 2.5 µl | Plasmid P19 template |
| 2.5 µl | 1:10 dilution of O15 (10 pmol/µL) |
| 2.5 µl | 1:10 dilution of O59 (10 pmol/µL) |
| 1.25 µl | dNTP mix |
| 35 µl | ddH2O |
| 1.25 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 29 | 95°C | 30 sec |
| 47 °C | 1 min | ||
| 68°C | 3 min | ||
| 3 | 1 | 68°C | 5 min |
| 4 | 1 | 4°C | infinity |
PCR successful - Band at about 2,1 kb :
Monday, July 16th
Preperative digestion and gelelectrophoresis of P98 and PCR26
Investigator: Jeff, Georg
Aim of the experiment: Construction of Gal4AD-Pif3, a part of the light-switchable promoter system.
Operational sequence:
Preparatory culture of S. cerevisiae
Investigator: Alois, Martin
Aim of the experiment: In order to have sufficient viable cells to inoculate an experimental culture in which we want to compare the growth rate of s. cerevisiae with a strain of brewing yeast (34/70 s. pastorianus weihenstephan) we aim to establish a preparatory over night culture.
Operational sequence: A sample of -80°C stored s. cerevisiae (glycerol stock) is used to inoculate 20ml of YPD-medium. This is shaken overnight at 30°C.
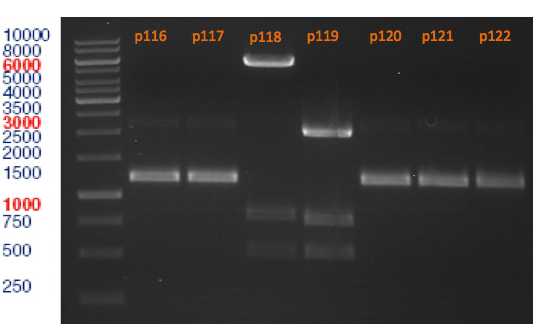
Miniprep of plasmids containing lavendula limonene synthase/citrus limonene synthase
Investigator: Lara
Aim of the experiment: Extract plasmids (pYESnew and pBS1C3) that contain limonene synthase/citrus limonene synthase.
Miniprep with Qiagen Kit; Plasmids p116-p122. Restriction digest with Xba1 and Pst1.
Restriction digest of p116-p122
Investigator: Lara
Aim of the experiment: Analytical restriction digest of plasmids p116, p117, p118, p119, p120, p121, p122.
Restriction digest with Xba1 and Pst1.
Analytical gel electrophoresis
Investigator: Lara
Aim of the experiment: Analytical gel electrophoresis of products from restriction digest of plasmids p116, p117, p118, p119, p120, p121, p122.
Midiprep of pSB1C3 (RFC25-compatible)
Investigator:Mary
Aim of the experiment: Get a lot of pSB1C3 for further experiments
Midipreparation of the pellet from over night cultures (E.coli, Transformed with pSB1C3 - RFC25)
was done with the Midiprep Kit from Qiagen
Result was named as P123 and had the concentration: 469,5 ng/µl (100µl at all)
PCR-Products of PAL (consless): digestion, extraction and ligation
investigator: Daniela, Mary
Aim of the experiment: digestion of PCR-Products to gain the correct cutting sites for ligation in pYES2 and pSB1C3 afterwards
Purification of PCR-Products with PCR-Purification Kit
- digestion with Xba1 and HF-Age1 (both NEB)
| volume | reagent |
| 25µl | PCR-product |
| 5µl | Buffer NEB4 |
| 0.5µl | BSA |
| 1µl | Xba1 (NEB; 20u/µl) |
| 1µl | HF-Age1 (NEB; 20u/µl) |
| 17.5µl | bidest. sterile H2O |
digestion took 3h at 37°C
- extraction from preparative gel:
PCR of PAL+cons using the optimized primer annealing temperature of 47 °C
Investigator: Daniela
Aim of the experiment: Amplification of the PAL cons gene contained in P19.
PCR
Reaction batch
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 2.5 µl | Plasmid P19 template |
| 2.5 µl | 1:10 dilution of O22 (10 pmol/µL) |
| 2.5 µl | 1:10 dilution of O59 (10 pmol/µL) |
| 1.25 µl | dNTP mix |
| 35 µl | ddH2O |
| 1.25 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 29 | 95°C | 30 sec |
| 47 °C | 1 min | ||
| 68°C | 3 min | ||
| 3 | 1 | 68°C | 5 min |
| 4 | 1 | 4°C | infinity |
PCR was succesful, band at 2,1kb:
Transformation of P94 - P97 (ligationproducts of pSB1C3 with CHS and OMT respectively) in E.coli
Investigator: Daniela
- adding 5µl ligation product (P94-P97) in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (without antibiotica) and incubate at 37°C, 45 min, 180 rpm
- plate on agar with chloramphenicol over night
-> were succesfull: some colonies were grown
Tuesday, July 17th
Preparative digest of P123 (pSB1C3 RFC25)
Investigator: Mary, Daniela
Digestion of P123 with Xbal/PstI and XbaI/AgeI
| volume | reagent |
| 23.1 µl | ddH2O |
| 4 µl | NEB4 |
| 0.4 µl | 10x BSA |
| 1 µl | XbaI (20 U/µl) |
| 1.5 µl | PstI (10U/µl) |
| 10 µl | P123 |
| volume | reagent |
| 23.35 µl | ddH2O |
| 4 µl | NEB4 |
| 0.4 µl | 10x BSA |
| 1 µl | XbaI (20 U/µl) |
| 1.25 µl | AgeI (10U/µl) |
| 10 µl | P123 |
Incubation: 37 °C, 3 h
DNA preparative gel electrophoresis
- gel: 1% with LMP-agarose
- band 1: P123 digested with XbaI and PstI
- band 2: P123 digested with XbaI and AgeI
- 70 V, 90 min
Gelextration
- cut the bands
- QIAquick Gel Extractrion Kit was used
- step 6 was left out
- step 9: 30µl buffer, 4 min incubation
Products were names as follows:
- P123 digested with XbaI and PstI: P132
- P123 digested with XbaI and AgeI: P133
PCR-Products of PAL (with consensussequence): digestion and extraction
investigator: Daniela, Mary
Aim of the experiment: digestion of PCR-Products to gain the correct cutting sites for ligation in pYES2 and pSB1C3 afterwards
Purification of PCR-Products with PCR-Purification Kit
- digestion with Xba1 and HF-Age1 (both NEB)
| volume | reagent |
| 25µl | PCR-product |
| 5µl | Buffer NEB4 |
| 0.5µl | BSA |
| 1µl | Xba1 (NEB; 20u/µl) |
| 1µl | HF-Age1 (NEB; 20u/µl) |
| 17.5µl | bidest. sterile H2O |
digestion took 3h at 37°C
- extraction from preparative gel:
Picking Clones of CHS and OMT in pSB1C3-RFC25
investigator: Daniela, Mary
Aim of the experiment: preculture over night for the miniprep next day
Repetition of ligation of PCR 15-20 (4CL, CHS and OMT) with pYES (P71 and P72 respectively)
Investigator: Mary, Daniela
Concentration (Nano Drop:
4CL+ (PCR 15) = 40.3 ng/µl
4CL- (PCR 16) = 37.3 ng/µl
CHS+ (PCR 17) = 46.1 ng/µl
CHS- (PCR 18) = 51.2 ng/µl
OMT+ (PCR 19) = 22.3 ng/µl
OMT- (PCR 20) = 16.7 ng/µl
P50 digested with XbaI and PstI = 23.9 ng/µl (the digested Plasmid has the number P71)
P50 digested with XbaI and NgOMIV = 28.4 ng/µl (the digested Plasmid has the number P72)
- required volumes were calculated using Lab Tools
4CL+ (PCR 15) with P71
| volume | reagent |
| 5.27 µl | P71 |
| 2.73 µl | 4CL+ (PCR 15) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
4CL- (PCR 16) with P71
| volume | reagent |
| 5.13 µl | P71 |
| 2.87 µl | 4CL- (PCR 16) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
CHS+ (PCR 17) with P72
| volume | reagent |
| 5.84 µl | P72 |
| 2.16 µl | CHS+ (PCR 17) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
CHS- (PCR 18) with P72
| volume | reagent |
| 6 µl | P72 |
| 2 µl | CHS- (PCR 18) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT+ (PCR 19) with P72
| volume | reagent |
| 4.45 µl | P72 |
| 3.55 µl | OMT+ (PCR 19) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT- (PCR 20) with P72
| volume | reagent |
| 3.87 µl | P72 |
| 4,13 µl | OMT- (PCR 20) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
Negative control
| volume | reagent |
| 5 µl | P71 or P72 |
| 3 µl | ddH2O |
| 1 µl | T4 DNA-ligase |
| 1 µl | T4-ligase buffer (10x) |
- water bath 16 °C
Transformation of P??- P??(ligationproducts of pYES with OMT and CHS respectively) in E.coli
- adding 5µl ligation product (P??- P??) in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (without antibiotica) and incubate at 37°C, 45 min, 180 rpm
- plate on agar with ampicillin over night
Inoculation of YPD medium with S. cerevisiae and brewing yeast strain 34/70 (part 1/3)
investigator: Maddin, Aloisius
Aim of the experiment: Discrimination of growing ability in wort medium
4 different worts:
- 21%, without hop
- 21%, without hop, autoclaved
- 16%, with hop
- 16%, with hop, autoclaved
were diluted with ELGA to a solution of 12%. These 4 different worts were inoculated with 5ml of preparatory culture of S. cerevisiae and 1ml of brewing yeast strain 34/70 (provided by the Forschungsbrauerei, Weihenstephan). Start OD was measured at 550nm (for table see next experiment). Those 8 flasks were incubated over night at room temperature and 130 rpm.
Wednesday, July 18th
Inoculation of YPD medium with S. cerevisiae and brewing yeast strain 34/70 (part 2/3)
Investigators: Martin, Alois
Aim of the experiment: Discrimination of growing ability in wort medium.
After 12h of cultivating the optical density (OD) at 550nm was determined:
| Assay | Yeast | Autoclaved | Hop | OD 0h | OD 12h | delta OD |
| A1 | S. cerevisae | No | No | 1.2 | 2.2 | + 1.0 |
| A2 | S. cerevisae | Yes | No | 2.0 | 2.6 | + 0.6 |
| A3 | S. cerevisae | No | Yes | 1.8 | 2.4 | + 0.6 |
| A4 | S. cerevisae | Yes | Yes | 2.4 | 2.7 | + 0.3 |
| B1 | 34/70 | No | No | 1.6 | 2.5 | + 0.9 |
| B2 | 34/70 | Yes | No | 3.0 | 2.9 | - 0.1 |
| B3 | 34/70 | No | Yes | 2.2 | 2.5 | + 0.3 |
| B4 | 34/70 | Yes | Yes | 2.7 | 2.8 | + 0.1 |
After 12h of cultivating the S. cerevisiae is growing quite well. There is a tendency that the brewing yeast strain 34/70 is not capable to grow sufficiently in autoclaved (e.g. proteinfree) medium. Since the 34/70 was stored at 4°C over 5 days, directly used to inoculate the medium (no preparatory culture) and we only used 1ml to inoculate with (compared to 5ml of the preparatory culture of S. cerevisiae - because of obvious differences in opacity of the inoculation media) there is not yet a conclusion to be drawn. We decided to incubate for another 24h at least.
new Miniprep of P50 pYES2
Investigators: Andrea
NanoDrop concentration: 303.6 ng/µl (260/280: 1.89)
Transformation of P50 (pYes) in E.coli
Investigator: Katrin
Aim of the experiment: Get more P50(pYes) for further experiments
- adding 2-3 µl of plasmid P50 in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (withouth antibiotica) and incubate at 37°C, 30 min, 180 rpm
- plate on agar with Ampicillin over night
 "
"