Team:Paris-Saclay/Project/Notebook/Week 13
From 2012.igem.org
Revision as of 14:45, 26 September 2012 by YohannPetiot (Talk | contribs)
27th August
| Visualization by electrophoresis of the amplification of 880bp end of BBa_K098995 by PCR from the 15 candidate colonies and #11. Use of a 0.8% Agarose gel. We are expecting a band at 880 bp | |
| Digestion by AseI of the 15 candidate colonies and #11. Visualization by electrophoresis on a 0.8% Agarose gel |
- Miniprep of colonies #11 and #21
- Plasmids of colonies #11 and #21 have been send for sequencing
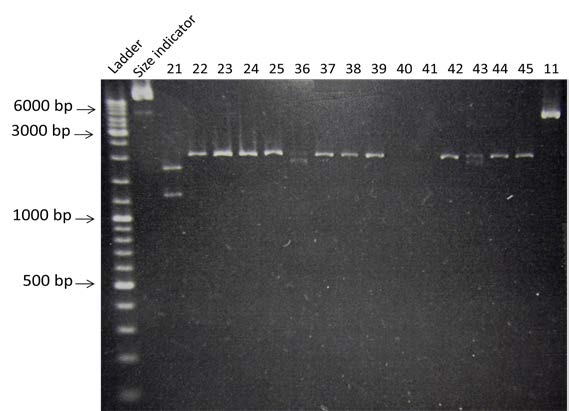
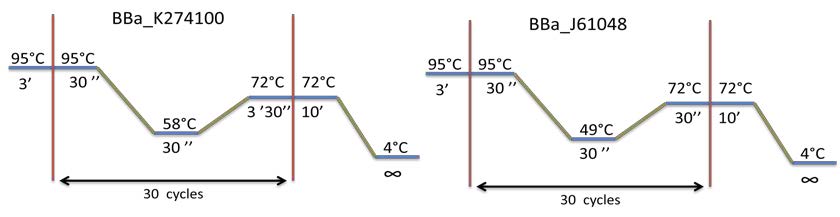
| Amplification of BBa_K274100, BBa_K115017 and BBa_J61048 by PCR using the colony #11. Visualization by electrophoresis on a 1% Agarose gel for BB3 and BB4 and a 2% Agarose gel for BB2. We are expecting a band at 123 bp for BBa_K115017, 133 bp for BBa_J61048 and 3408bp for BBa_K274100. |
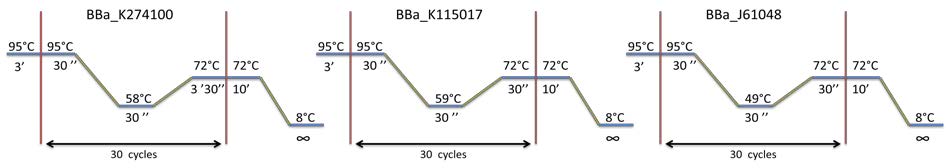
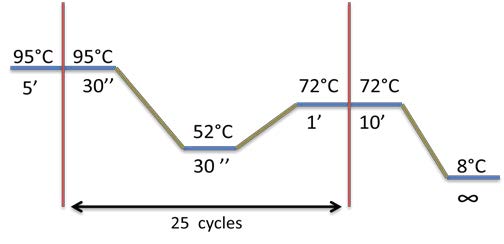
PCR program used for each biobrick amplification:
- Gibson assembly for the A construction (BBa_K098995+BBa_K274100+ BBa_J61048+Plasmid pSB1A2) and for the C construction (BBa_K115017 + BBa_C0051+166bp end of BBa_K098995+BBa_K274100+ BBa_J61048+Plasmid pSB1A2). Transformation of DH5αZ1 competent cell with construction A or construction C. Petri Dishes have been placed at 37°C for 1 day (construction A) or 2 days (construction C)
28th august
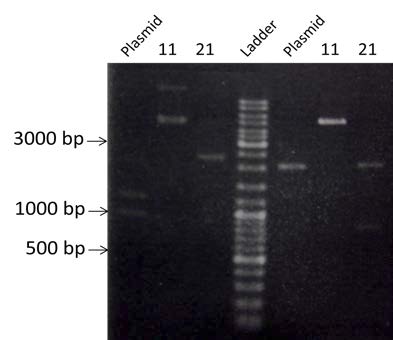
Digestion by AseI and NotI of the colonies #11, #21 and the linear plasmid.
- AseI:
- 1 restriction site in the Plasmid pSB1A2
- 1 restriction site in the 880bp end of BBa_K098995
- NotI:
- 1 restriction site either side of the B construction
| Visualization by electrophoresis on a 0.8% Agarose gel | |
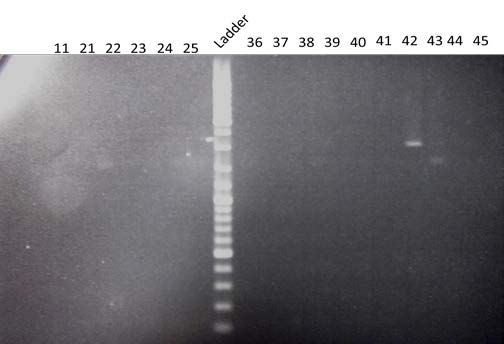
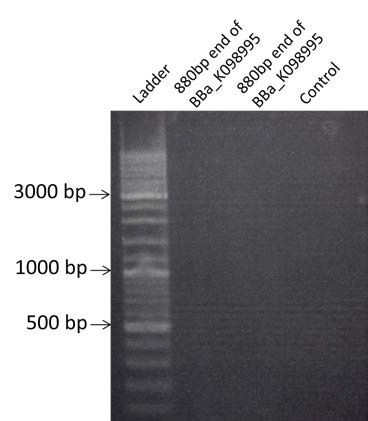
| Amplification by PCR of the 880bp end of BBa_K098995. Visualization by electrophoresis on a 0.8% Agarose gel. We are expecting a band at 880bp. |
PCR program used:
- Amplification by PCR of the 880bp end of BBa_K098995 + BBa_K115017 contained in the colonies #11 and #21. PCR program used:
- Miniprep of the colony #11.
- Gibson assembly of the construction B (BBa_K115017+880bp end of BBa_K098995+ BBa_K274100+ BBa_J61048+ pSB1A2). Transformation of competent cells DH5α with the construction.
- Liquid culture of the colony #11 and a control colony 11811 to prepare a test of temperature range.
| Digestion of the colony #11 by AseI+HindIII. Visualization by electrophoresis on a 0.8% Agarose gel. |
29th August
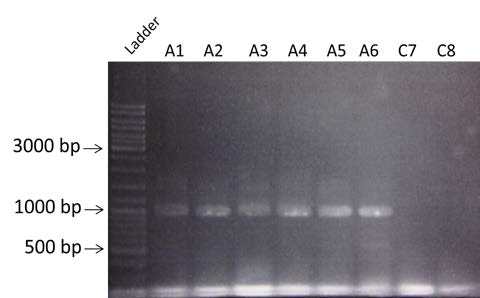
| PCR on colonies to verify the construction A and C obtained by Gibson assembly. Amplification of BBa_K098995 for A and BBa_C0051 for C. Visualization by electrophoresis on a 0.8% Agarose gel. We are expecting a band at 935bp for A and 750pb for C. |
- Miniprep of the construction A and the construction C.
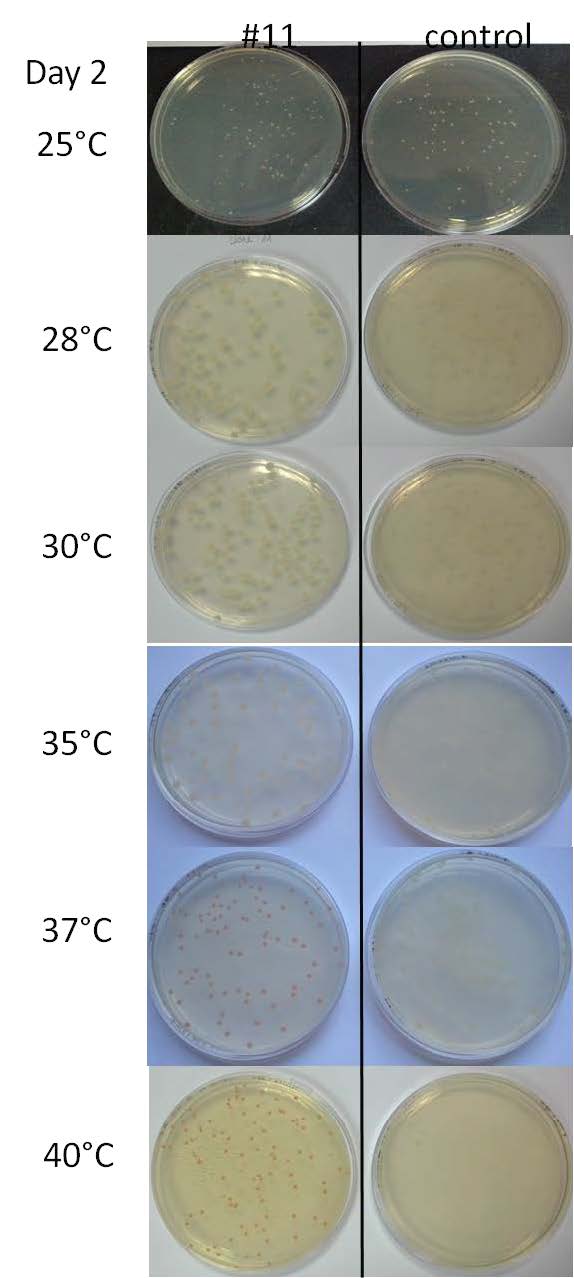
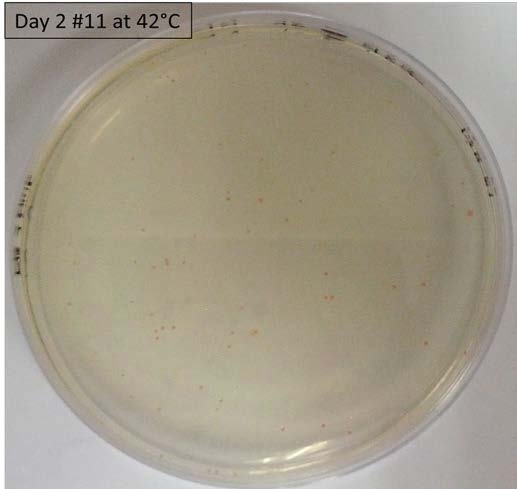
- Performing tests on a range of temperatures with the liquid culture of the colony #11 and the control 11811. On Petri Dishes LB+ agar+Ampicilline. 7 different temperatures have been tested: 25°C, 28°C, 30°C, 35°C, 37°C, 40°C and 42°C.
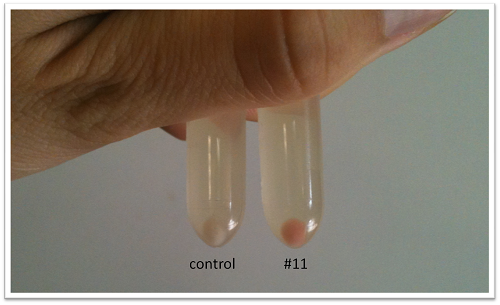
- The remaining of liquid cultures has been centrifuged to see the residue.
30th and 31th august
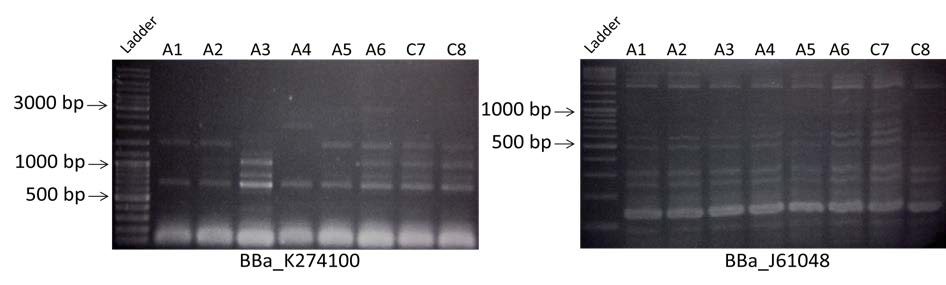
- Amplification of BBa_K274100 and BBa_J61048 of the colonies containing the construction A or the construction C. Visualization by electrophoresis on a 0.8% Agarose gel for BBa_K274100 and a 2% Agarose gel for BBa_J61048.
PCR program used:
- Glycerol stock of the colonies #11 and #21.
 "
"






Follow us !