Team:NRP-UEA-Norwich/NOSensing
From 2012.igem.org
Welcome to the NRP UEA iGEM 2012 Wiki Projects Menu
Please choose the relevant link to view an overview of each project!
Nitric Oxide Sensing & The Hybrid Promoters | The Comparator Circuit | Theoretical Projects
Our main project has resulted in the production of a hybrid bacterial and mammalian promoter optimised for induction by nitric oxide, nitrates and nitrites. We have ligated PyeaR, a known bacterial promoter and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K216005 Part BBa_K216005] in the parts registry, with its mammalian counterpart, CArG. The resulting hybrid promoter has been synthesised in two orientations; PyeaR (bacterial, B) upstream of CArG (mammalian, M), nicknamed (B-M); and CArG upstream of PyeaR (M-B). These orientations were submitted to the parts registry as our first two biobricks.
Each orientation of the promoter was ligated to enhanced cyan fluorescent protein (eCFP) and red fluorescent protein (RFP) to produce four new biobricks which have been submitted to the parts registry. These promoter + fluorescent protein biobricks have been characterised following transformation into Escherichia coli and induction by potassium nitrate using methods such as flow cytometry, fluorescence-activated cell sorting (FACS) and scanning with a fluorometer. The data from these experiments has proved that our promoter works as we expected it to. We have also transfected M-B + eCFP into a human breast cancer cell line, MCF7, and have proved the system is flexible and can be used in both eukaryotes and prokaryotes.
We believe the promoters we have produced have relevant uses in cancer therapeutics, soil fertilisation and detection of emissions from industries such as construction.
Parts produced from this project:
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K774000 Bacterial-Mammalian/B-M (PyeaR-CArG) Hybrid Promoter] -- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K774001 Mammalian-Bacterial/M-B (CArG-PyeaR) Hybrid Promoter] -- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K774004 B-M + eCFP] -- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K774005 B-M + RFP] -- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K774006 M-B + eCFP] -- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K774004 M-B + RFP]
The University of East Anglia hosts many research teams whose work focuses on studying nitrogenous species and the way in which bacteria use and modify those species. One major problem that is faced by these teams involves working with nitric oxide (NO), as NO is highly reactive with a low half life, therefore making it difficult to trace and quantitatively measure accurately. Many of the methods currently used to measure NO levels are unable to distinguish between homogenous species, such as nitrates (NO3) and nitrites (NO2), therefore the figure given for NO levels is often inaccurate as other nitrogenous species are taken into account.
The ability to be able to accurately detect NO levels is one with a great deal of potential for the future. Nitric oxide has been noted as a possible cancer therapy due to its physiological use as an apoptosis inducer by macrophages, however NO is also known to be used by cancerous cells to establish a baseline and use it to induce apoptosis and promote proliferation of a tumour; being able to accurately sense nitric oxide and go on to act on that information could be very useful to prevent the NO baseline being established by cancerous cells, but to also use NO for its apoptosis-inducing abilities. There are also other potential applications in the construction business, in 2008 NEED TO FIND REFERENCE OF PEOPLE released legislation encouraging construction companies to monitor their NO output as it is inversely proportional to carbon monoxide (a toxic substance which needs to be regulated); the ability to accurately detail levels of NO being released in these circumstances would be highly useful.
Nitric oxide is an extremely physiologically relevant molecule found within both eukaryotes and prokaryotes, where it is utilised by different enzymes and systems for various roles. The aim of the experiment was to devise a hybrid promoter that could be applied to eukaryotes and prokaryotes in order to begin to more accurately sense NO and report on its specific levels.
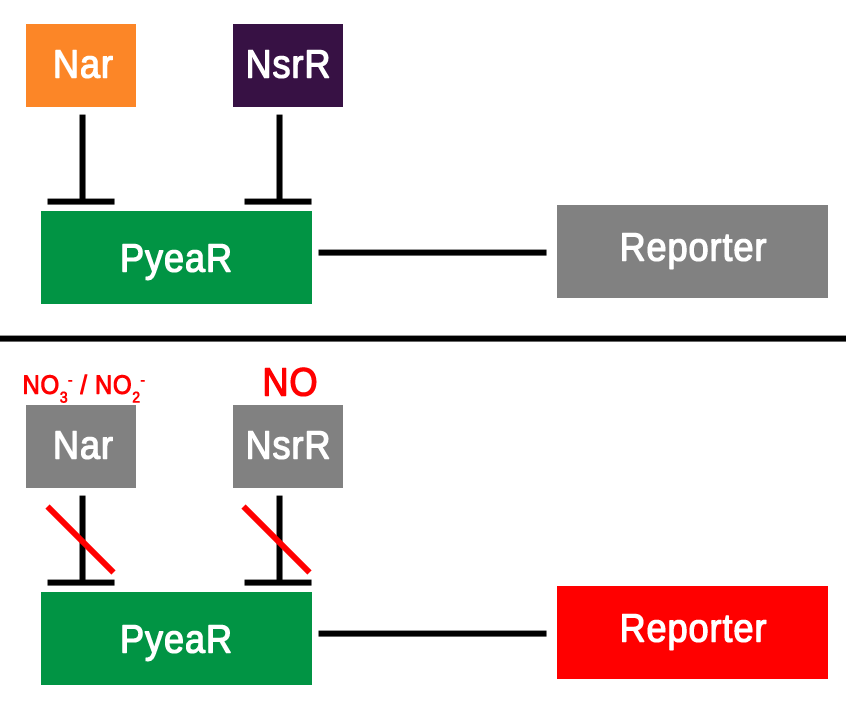
The hybrid promoter was designed to take on both bacterial and mammalian elements in order to be compatible with both bacterial and mammalian cells. After research around the subject and searching the parts registry a promoter known as PyeaR was decided upon as the bacterial element of the hybrid promoter; PyeaR is found in the yeaR/yoaG operon of Escherichia coli and is associated with induction by nitric oxide, nitrates and nitrites (Lin et al., 2007). PyeaR is repressed by two main repressors; Nar, which is regulated by nitrates and nitrites; and NsrR, which is regulated by nitric oxide (Figure 1.). One of the key elements of PyeaR is that it is not repressed in aerobic conditions, allowing for easier carrying out of experiments. The PyeaR aspect of the hybrid promoter has been known throughout the project as the bacterial promoter, or simply B.
The mammalian element of the hybrid promoter was produced by nine CArG elements (repeated elements of CC(A/T)(6)GG), a promoter previously used synthetically for nitric oxide synthase as a cancer therapy (Worthington et al., 2005) and developed from the EGR1 gene for early growth response protein 1 (Scott et al., 2002). The CArG aspect of the hybrid promoter has been known throughout the project as the mammalian promoter, or simply M.
Following identification of the two elements of the hybrid promoters the B (PyeaR) and M (CArG) aspects were ligated to one another in two orientations; B upstream of M (B-M) and M upstream of M (M-B) (Figure 2.). The hybrid promoters were synthesised in a pUC57 backbone with the standard iGEM restriction sites of EcoR1/Xba1 upstream of the promoter, and Spe1/Pst1 downstream of the promoter. A BamH1 restriction site was included in between the B and M sequences in order to allow for the B and M elements to be separated, as well as for easy verification of the promoter having been ligated into the iGEM backbone in future experiments (as BamH1 does not already exist in the pSB1C3 backbone).
The DNA for the synthesised genes of B-M and M-B had been supplied in the pUC57 backbone, therefore it was necessary for B-M and M-B to be digested from the pUC57 backbone and ligated into the pSB1C3 backbone. The synthesised gene was transformed into competent ’’E. coli’’ cells, which in turn were grown on agar plates containing 100 µg/ml ampicillin (due to pUC57 containing ampicillin resistance); colonies that had grown were then inoculated into liquid culture, and the liquid culture was subsequently mini-prepped using either the Bioline ISOLATE Plasmid DNA Mini Kit or the Promega Wizard® Plus SV Minipreps DNA Purification System. The DNA that had been extracted through mini-preps and the pSB1C3 backbone, as provided by the iGEM registry, were then digested using EcoR1 and Pst1 and a ligation was carried out using standard assembly protocol. The product of ligation was then transformed into competent E. coli cells, which were grown on agar plates containing 2.5 µg/ml chloramphenicol (due to pSB1C3 containing chloramphenicol resistance); this was done to eliminate any bacteria that had been transformed with undesirable ligation products.
The colonies that had grown were then grown in liquid culture and mini-prepped in order to extract the DNA; the extracted DNA was then sent for sequencing, and the returned sequenced matched the expected sequence. The DNA was then sent to the parts registry as the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K774000 Bacterial-Mammalian Hybrid Promoter] and the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K774001 Mammalian-Bacterial Hybrid Promoter]. The biobricks for B-M and M-B were then used for further experiments, including ligation with a fluorescent protein reporter and growth studies.
From weeks one through to five the team worked on producing the first biobricks of the hybrid promoter. Despite this proving difficult due to various reasons such as low amounts of DNA being produced from early mini-preps and ligation strategies not working, E. coli transformed by the biobrick DNA was successfully grown and proven to have the relevant resistance by the beginning of week six.
In order to test the activity of the hybrid promoters a reporter needed to be ligated. As the hybrid promoter did not already contain a ribosome binding site (RBS) both the RBS and the reporter were needed to be ligated to the promoter; in order to help improve experimental efficiency the parts registry was searched for relevant reporters that also contained an RBS. In week three two reporters were identified as [http://partsregistry.org/Part:BBa_E0420 BBa_E0420], a biobrick for enhanced CFP (eCFP) + RBS + terminators, and [http://partsregistry.org/Part:BBa_K081014 BBa_K081014], a biobrick for RFP + RBS + terminators.
Once the B-M and M-B biobricks had been created in week six work began in earnest on the fluorescent proteins and ligating the promoters to them in order to begin characterisation. Due to many set-backs with low levels of DNA and having to order more biobricks from the registry, a successful ligation of the promoter to a fluorescent protein reporter was finally achieved in week ten. In order to carry out the ligation the promoter was first digested using Spe1 and Pst1 in order to linearise the backbone downstream of the promoter; the fluorescent proteins were digested using Xba1 and Pst1 in order to remove the insert. A ligation was then carried out using standard assembly protocol and the ligation products were transformed into E. coli competent cells, which in turn were grown on agar plates 2.5 µg/ml chloramphenicol (due to pSB1C3 containing chloramphenicol resistance).
. Used RFP/CFP with the fluorometer etc. to get data on sensitivity
. Tranfected into mammalian cells to show flexibility
. Studied the growth rate
. Photos
. Graphs from flow and fluorometer
. Pictures etc. from mammalian transfection
. Growth study on MB/BM
BM/MB better or the same, RFP/CFP better or the same etc.
A problem with current cancer therapeutic techniques is that they are not specific to cancer cells, leading to patient discomfort. We hope to engineer E.coli cells that selectively identifies cancerous cells through their anaerobic environment and secrete high concentrations of NO, mimicking macrophages, and thus killing these cells. These cells are known to be hypoxic, and hypoxia is a hallmark for cancer cells already used in tumour detection. Our Warrior Cell construct hopes to pave the way for a new kind of therapeutic strategy that delivers a therapeutic agent only when and where it is needed.
. Full quantitative analysis to see where the values lie; combine with tuners for different sensitivity levels
. With different substrates (e.g. nitrite salt, NO donor)
. With different reporter/effector enzyme
Lin H.Y., Bledsoe P.J., Stewart V., (2007), Activation of yeaR-yoaG Operon Transcription by the Nitrate-Responsive Regulator NarL Is Independent of Oxygen- Responsive Regulator Fnr in Escherichia coli K-12▿, Journal of Bacteriology, 189: 7539 - 7548
Scott S.D., Joiner M.C., Marples B., (2002), Optimizing radiation-responsive gene promoters for radiogenetic cancer therapy., Gene Therapy, 9: 1396-1402
Worthington J., Robson T., Scott S., Hirst, D., (2005), Evaluation of a synthetic CArG promoter for nitric oxide synthase gene therapy of cancer, Gene Therapy, 12: 1417–1423