Team:Kyoto/Secretion/Notebook
From 2012.igem.org
Secretion Notebook
February 7
Preculture
We started preculture at 12:10.
February 8
Culture
We start culturing with 300[mL] of LB medium.
| time | OD600 |
|---|---|
| 12:00 | start |
| 14:10 | 0.019 |
| 14:45 | 0.154 |
| 15:05 | 0.267 |
| 15:21 | 0.64 |
Making Competent Cell
We made competent cells.
Transformation
pGEM_TAP
Lacp (BBa_R0011)
DT (BBa_B0015)
Making Culture Medium Plates
We made 200mL of ampicillin culture, kanamycin culture, and chloramphenicol culture.
Transformation
GFP(BBa_E0040) in pSB1A2
DT(BBa_B0015) in pSB1AK3
ara(BBa_I0500) in pSB2K3
Lacp(BBa_R0011) in pSB1A2
February 9
transformation
BBa-E0040(GFP)(Mr.Fujita)
Liquid culture
DT.leap colony transformed at feb.8
competent cell onfeb.8
February 10
Miniprep
DT2 43.9μg/ml(1.34 260/230 1.74 260/280)
lacp1 17.1μg/ml(1.54 260/280 0.83 260/230)
lacp2 18.0μg/ml(1.58 260/280 0.87 260/230)
transformation
B0040 1.4k PsB1A2 B0034 1.2M pSB1A2(from iGEM parts plate)
Competent cell
We did preculture for overnight. We put 1.5mL of preculture on 150mL of LB culture.
| time | OD600 |
|---|---|
| 11:45 | start |
| 13:30 | 0.048 |
| 14:30 | 0.168 |
| 15:03 | 0.256 |
| 15:20 | 0.405 |
| 15:35 | 0.459 |
| at last | 0.576 |
February 11
Checking Transformation efficiency
Cpnpetent cell's transformation efficiency is 1.3x10^4colonys/μg
February 13
Transformation
Const promoter J23110,J23109,J23100
| DNA | Competent cell | total |
|---|---|---|
| 1μL | 20 | 21 |
No colony was there on feb.14
Liquid culture
lacP,DT,RBS(BBa_B0034),GFP
start at 20:00
in Plus grow with Ampicilin 3mL
February 14
Miniprep
concentration[μg/μL]
| lacP3 | 39.6 |
| lacP4 | 40.8 |
| lacP5 | 28.9 |
| RBS1 | 28.2 |
| RBS2 | 57.4 |
| RBS3 | 13.2 |
| DT3 | 69.7 |
| DT4 | 64.4 |
| DT5 | 61.5 |
| GFP1 | 64.0 |
| GFP2 | 50.5 |
| GFP3 | 66.0 |
Restriction
Const promoterJ23100
| DNA | Spe1 | Pst1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for overnight
February 15
Making gel
1% Agarose gel
| Agarose | TAE |
|---|---|
| 1.6g | 160mL |
Electrophoresis
| Restriction product | loading dye |
|---|---|
| 5μL | 1 |
The marker was 1kb ladder
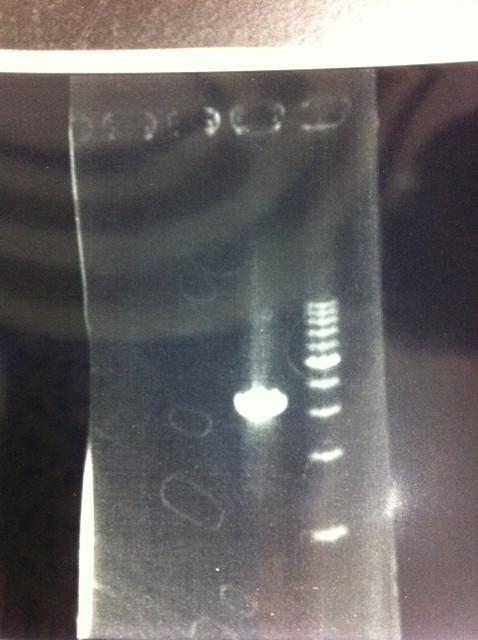
It seemed that this restriction product was not cut.
Lane1 : 1kb ladder
Lane2 : J23100 2μL + 6*Loading dye 1μL
Lane3 : J23100(Spe1,Pst1) 5μL
Lane4 : J23100(Spe1,Pst1) 2μL
- There were bands on lane_2 and we cannot identify these bands because the sample of lane_2 was not cut with any restriction enzyme.
- There must have been bands at 2100bp and 883bp on lane_3 and lane_4.
Testing whether restriction enzyme were deactivated or not
| DNA(DT) | restriction enzyme | Buffer | BSA | MilliQ | total |
|---|---|---|---|---|---|
| 10 | 0.5 | 3 | 0.5 | 16 | 30 |
at 37℃ for Oveernight
Restriction enzyme means Spe1(1,2) Pst1(1,2,3) in this time.
February 16
Electrophoresis
1. 1kb ladder
2. DT2
3. DT3
4. DT2 (Spe1-1)
5. DT2 (Spe1-2)
6. DT2 (Pst1-1)
7. DT2 (Pst1-2)
8. DT2 (Pst1-3)
9. DT3 (Pst1-4)
10. 1kb ladder
Pst1, Pst2, and Pst3 did not cut DNAs. They seemed to be deactivated.
Genomic PCR
| 10*Buffer for KOD Plus | 2mM dNTPs | 25mM MgSO4 | 10μM primer-f | 10μM primer-r | 158ng/μL Genomic DNA | KOD plus | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 1 | 32 | 50 |
Electrophoresis
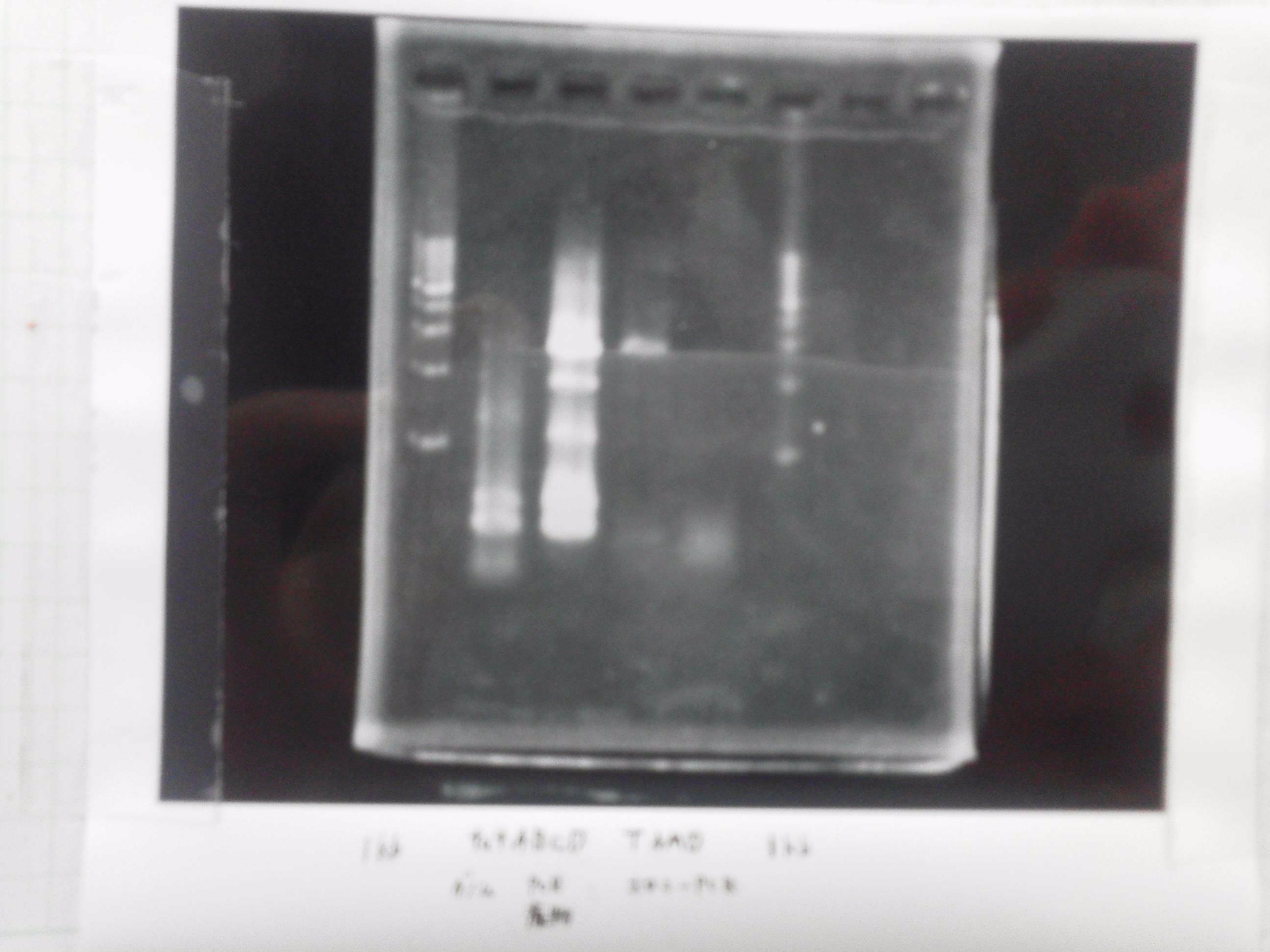
1. 1kb ladder
2. tatABCD (2.5kb)
3. TAMO reductase (2.7kb)
4. Negative control
We got bands of tatABCD but there were nonspecific amplification products.
We failed amplification of TAMO reductase.
Transformation
Constitutive promoter (BBa_J23107 , BBA_J23117)
High copy plasmid (pSB1AT3)
| DNA | competent cell |
|---|---|
| 1μL | 10μL |
February 17
PCR
We did PCR to amplify products of PCR that we had done yesterday but we could not amplify tatABCD.
Genomic PCR
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | genomic DNA | KOD plus | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 5 | 0.75 | 0.75 | 0.5 | 0.5 | 16 | 50 |
Predenature 94℃, 2min
Denature 98℃, 10sec
Annealing 57℃, 30sec
Extension 68℃, 2.5min
(30cycles)
Electrophoresis
1. 1kb ladder
2. TAMO reductase
3. Negative control
Restriction
| J23100 | Spe1 | Pst1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
Genomic PCR
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | genomic DNA | KOD plus | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| TMAO reductase | 2.5 | 2.5 | 3 | 0.75 | 0.75 | 0.5 | 0.5 | 15.5 | 25 |
| tatABCD | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 0.5 | 0.5 | 15.5 | 25 |
| tatABCD | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 5 | 0.5 | 10.5 | 25 |
Electrophoresis
1. 1kb ladder
2.3. TAMO reductase
4. tatABCD
5. tatABCD(10 times amount of genome)
Checking of restriction enzyme
| DT | enzyme | Buffer | BSA | MilliQ | total |
|---|---|---|---|---|---|
| 2 | 0.5 | 3 | 0.5 | 24 | 30 |
at 37℃ for overnight
We checked EcoR1 and Xba1.
February 18
Miniprep
| μg/mL | 260/280 | 230/260 | |
|---|---|---|---|
| JS3117-1 | 135 | 1.5 | 2.06 |
| JS3117-2 | 75 | 1.6 | 1.63 |
| JS3109-1 | 115 | 1.5 | 1.88 |
| JS3109-2 | 75 | 1.65 | 1.71 |
| pSBIAT3-1 | 70 | 1.66 | 1.83 |
| pSBIAT3-2 | 100 | 1.52 | 1.54 |
diluted to 25 times
Liquid culture
Competent cell
We put 3mL of preculture product on yesterday onto 300mL of LB medium
| time | OD600 |
|---|---|
| 10:30 | start |
| 12:10 | 0.118 |
| 13:00 | 0.270 |
| 13:30 | 0.502 |
Transformation
| pSB1AT3-2 | competent cell | MilliQ | total |
|---|---|---|---|
| 0 | 20 | 10 | 30 |
| 2 | 20 | 8 | 30 |
| 10 | 20 | 0 | 30 |
- Results(2/19)
number of colony 0 177 590
Transformation efficiency 7.4x10^4 colonys/μg
February 20
Restriction
sample 1
| DT plasmid | EcoR1 | Xba1 | Buffer | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 7.5 | 0.5 | 0.5 | 3 | 0.5 | 18 | 30 |
sample2
| GFP plasmid | EcoR1 | Spe1 | Buffer | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
Electrophoresis
- sample1
a. sample1 2μL + MilliQ 3μL + 6×Loading Dye 1μL
b. sample1 5μL + 6×Loading Dye 1μL
lane_1 1kb ladder
lane_2 a
lane_3 b
lane_4 a
lane_5 b
lane_6 a
lane_7 b
lane_8 1kb ladder
- sample2
c. sample2 2μL + MilliQ 3μL + 6×Loading Dye 1μL
d. sample2 5μL + 6×Loading Dye 1μL
lane_1 1kb ladder
lane_2 c
lane_3 d
lane_4 1kb ladder
PCR
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | genomic DNA | KOD plus | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| tatABCD1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 0.5 | 16 | 25 |
| tatABCD2 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 0.5 | 16 | 25 |
| TMAO reductase1 | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 0.5 | 0.5 | 15.5 | 25 |
| TMAO reductase2 | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 5 | 0.5 | 10.5 | 25 |
Predenature 94℃ 2min
Denature 98℃ 10sec
Annealing 59℃ 30sec
Extension 68℃ 2.5min
→30cycles
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | PCR products | genomic DNA | KOD plus | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|---|
| tatABCD1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0 | 0.5 | 0.5 | 16 | 25 |
| tatABCD2 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 1 | 0 | 0.5 | 15.5 | 25 |
| TMAO reductase1 | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 0 | 0 | 0.5 | 16 | 25 |
| TMAO reductase2 | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 0 | 0 | 0.5 | 16 | 25 |
Predenature 94℃ 2min
Denature 98℃ 10sec
Annealing 59℃ 30sec
Extension 68℃ 2.5min
→35cycles
Electrophoresis
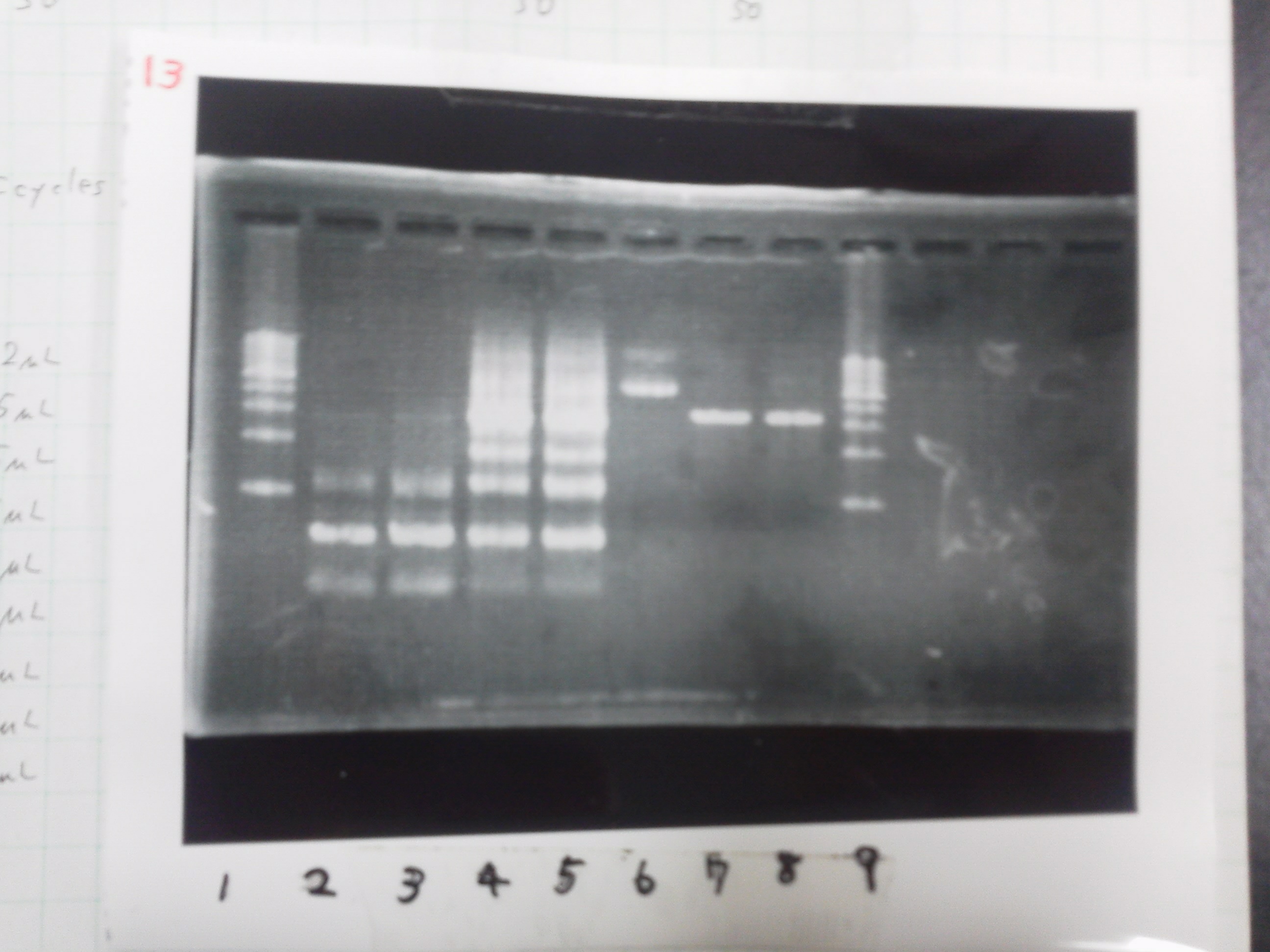
lane_1.1kb ladder
lane_2.tatABCD
lane_3.tatABCD
lane_4.TAMO1
lane_5.TAMO2
lane_6.1kb ladder
lane_1.1kb ladder
lane_2.tatABCD1
lane_3.tatABCD2
lane_4.TAMO
lane_5.TAMO
lane_6.1kb ladder
February 21
PCR (Advantage HF protocol)
| buffer | dNTPs | primer-f | primer-r | gDNA | PCR products | DW | polymerase | total | |
|---|---|---|---|---|---|---|---|---|---|
| tatABCD | 2.5 | 2.5 | 0.75 | 0.75 | 1 | 0 | 17 | 0.5 | 25 |
| TMAO | 2.5 | 2.5 | 0.75 | 0.75 | 0 | 1 | 17 | 0.5 | 25 |
Predenature 94℃ 1min
Denature 94℃ 30sec
Annealing 58℃ 30sec
Extension 68℃ 3min
→25cycles
Electrophoresis
1. 1kb ladder 2μL
2. tatABCD 5μL + 6×Loading Buffer 1μL
3. TAMO 5μL + 6×Loading Buffer 1μL
4. 1kb ladder 2μL
Liquid culture
pSB3C5-1,2
pSB4K5-1,2
February 22
Gel extraction
lane 1 of the gel 45.0μg/mL
PCR purification
product 38.2μg/mL
PCR
TMAO reductase
| Buffer | dNTPs | MgSO4 | prefix primer-f | suffix primer-r | product of gel extract | product of PCR purification(1ng/μL) | KOD plus | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | 4 | 1.5 | 1.5 | 0.5 | 0 | 1 | 32.5 | 50 |
| 2 | 5 | 5 | 4 | 1.5 | 1.5 | 1 | 0 | 1 | 32.5 | 50 |
| 3 | 5 | 5 | 4 | 1.5 | 1.5 | 0 | 0.5 | 1 | 32.5 | 50 |
| 4 | 5 | 5 | 4 | 1.5 | 1.5 | 0 | 1 | 1 | 32.5 | 50 |
94℃, 2min
98℃, 10sec
59℃, 30sec
68℃, 3min
→25cycles
Electrophoresis
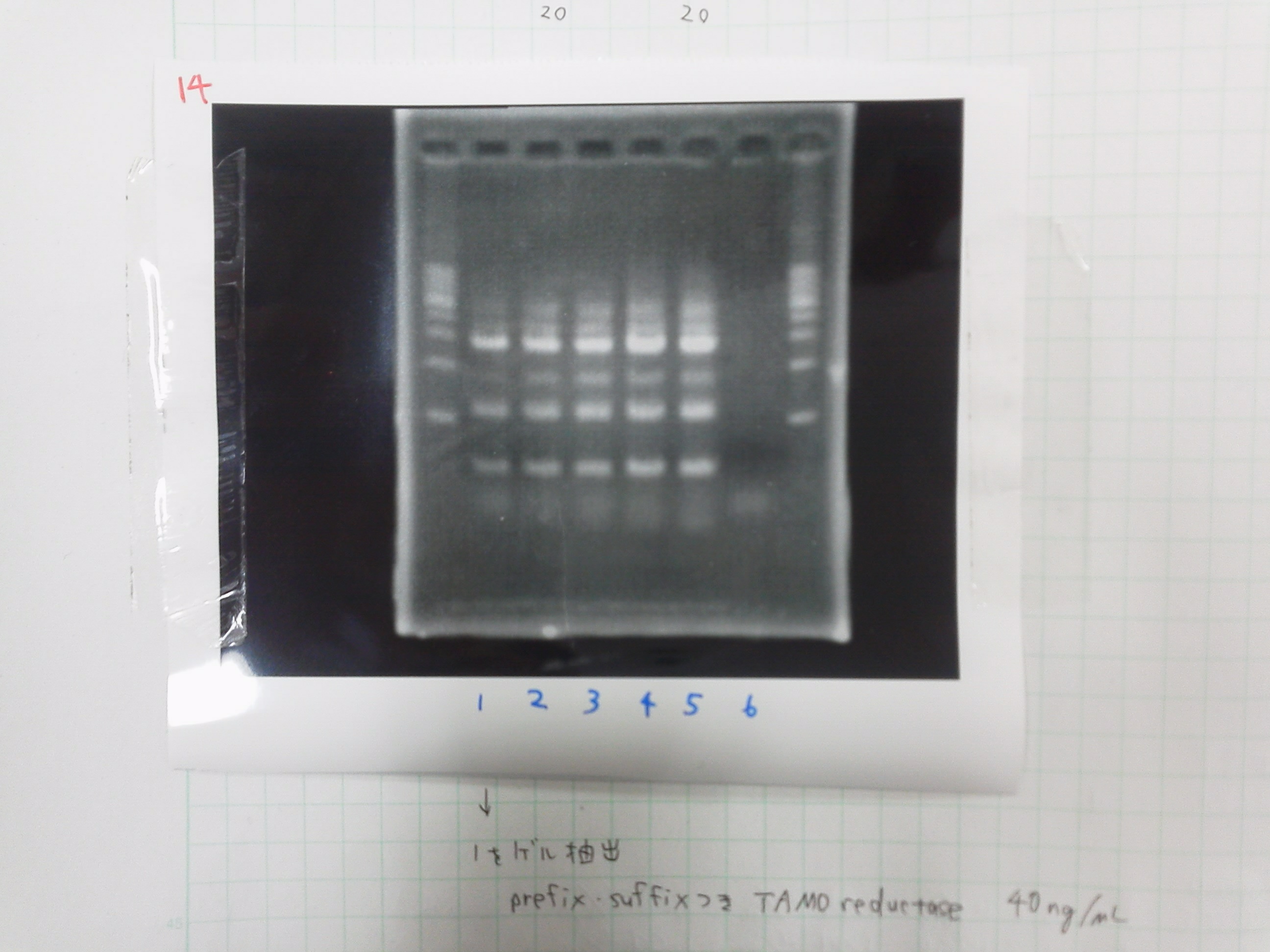
1. 1kb ladder
2. TAMO1
3. TAMO2
4. TAMO3
5. TAMO4
6. constructive promoter 1-18C
7. constructive promoter Spe1
8. constructive promoter Pst1
9. 1kb ladder
PCR
| Buffer | dNTPs | MgSO4 | prefix primer-f | suffix primer-r | product of PCR purification(1ng/μL) | KOD plus | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | 3 | 1.5 | 1.5 | 0.5 | 1 | 32.5 | 50 |
| 2 | 5 | 5 | 3 | 1.5 | 1.5 | 1 | 1 | 32 | 50 |
| 3 | 5 | 5 | 3 | 1.5 | 1.5 | 2 | 1 | 31 | 50 |
| 4 | 5 | 5 | 3 | 1.5 | 1.5 | 3 | 1 | 30 | 50 |
| 5 | 5 | 5 | 3 | 1.5 | 1.5 | 10 | 1 | 29 | 50 |
| 6 | 5 | 5 | 3 | 1.5 | 1.5 | 0 | 1 | 33 | 50 |
94℃, 2min
98℃, 10sec
59℃, 30sec
68℃, 3min
→25cycles
Electrophoresis
Checking Dpn1
| Buffer | lacp(28.7ng/μL) | Dpn1 | MilliQ | total |
|---|---|---|---|---|
| 2 | 10 | 0.5 | 7.5 | 20 |
| 2 | 10 | 0 | 5 | 17 |
February 23
Colony PCR
tatABCD(2 samples)
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | KOD plus | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
Predenature 94℃ 1min
Denature 98℃ 10sec
Annealing 59℃ 30sec
Extension 68℃ 3min
→25cycles
Electrophoresis
1. 1kb ladder 2μL
2. tatABCD 1 5μL + 6×Loading Buffer 1μL
3. tatABCD 2 5μL + 6×Loading Buffer 1μL
4. 1kb ladder 2μL
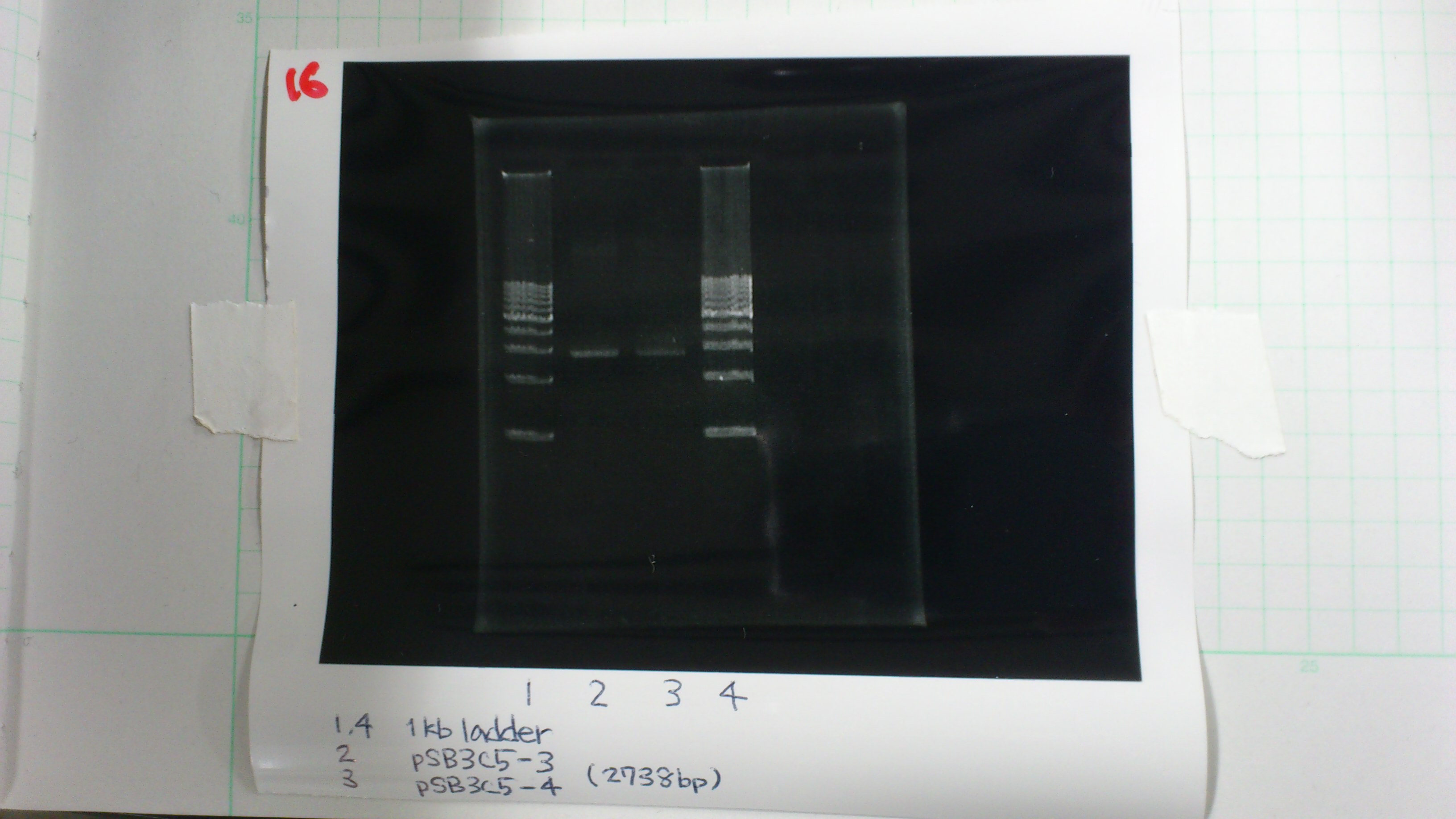
Miniprep
pSB4K5 and pSB3C5
deluted it to 25 times and then measured it
pSB4K5 1 : 60.0 μg/ml 1.67(260/280) 1.98(260/230)
pSB4K5 2 : 55.0 μg/ml 1.49(260/280) 1.62(260/230)
pSB3C5-3 : 3.6 μg/ml 1.57(260/280) 3.00(260/230)
pSB3C5-4 : 1.3 μg/ml 1.44(260/280) 1.04(260/230)
Liquid culture
pSB3C5-3,4
Electrophoresis
1. 1kb ladder 2μL
2. pSB3C5-3 5μL, 6×loading dye 1μL
3. pSB3C5-4 5μL, 6×loading dye 1μL
4. 1kb ladder 2μL
February 27
Test of Dpn1
| Buffer2 | GFP2 | BSA | MilliQ | Dpn1 |
|---|---|---|---|---|
| 3 | 3 | 0.3 | 23 | 1 |
Colony PCR
| buffer | dNTPs | MgSO4 | Primer-f | Primer-r | MilliQ | KOD Plus | total | |
|---|---|---|---|---|---|---|---|---|
| Colony PCR(2 samples) | 5 | 5 | 3 | 1.5 | 1.5 | 33 | 1 | 50 |
| Negative control | 5 | 5 | 3 | 1.5 | 1.5 | 34 | 0 | 50 |
Predenature 94℃,2min
Denature 98℃,10sec
Annealing 59℃,30sec
Extension 68℃,3min
→25cycles
Electrophoresis
| sample | Loading Dye | MilliQ | |
|---|---|---|---|
| 1.1kb ladder | 2 | 0 | 0 |
| 2.product of PCR1 | 5 | 1 | 0 |
| 3.product of PCR2 | 5 | 1 | 0 |
| 4.product of PCR(Negative control) | 5 | 1 | 0 |
| 5.product of PCR(2/23) | 5 | 1 | 0 |
| 6.GFP2(DPN1) | 10 | 2 | 0 |
| 7.GFP2 | 3 | 2 | 7 |
| 8.1kb ladder | 2 | 0 | 0 |
Results of liquid culture
We measure this after dilute it to 10 times.
| pSB3C5-5 | pSB3C5-6 | pSB3C5-5(1% glucose) | pSB3C5-6(1% glucose) |
|---|---|---|---|
| 8.5[µg/ml] | -1.8 | -17.9 | -18.2 |
PCR
| buffer | dNTPs | MgSO4 | Primer-f(prefix) | Primer-r(suffix) | PCR purification product(1ng/µL) | MilliQ | KOD Plus | total | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | 3 | 1.5 | 1.5 | 0.2 | 32.8 | 1 | 50 |
| 2 | 5 | 5 | 3 | 1.5 | 1.5 | 0.5 | 32.5 | 1 | 50 |
- PCR purification product was that purification product(75ng/µL) of electrophoresis-3 deluted to 1ng/µL
Predenature 94℃,2min
Denature 98℃,10sec
Annealing 59℃,30sec
Extension 68℃,3min
→25cycles
February 28
Electrophoresis
1. 1kb ladder
2. PCR1 →Product of gel extraction : tatABCD with prefix and suffix 105[ng/µL]
3. PCR2
Restriction
| Buffer2 | plasmid(?) | enzyme | MilliQ | total |
|---|---|---|---|---|
| 2 | 2 | 0.2 | 15.8 | 20 |
incubate 1 hour at 37℃
Electrophoresis
1. 1kb ladder
2. Control (without enzymes)
3. EcoR1
4. Xba1 (crystallized)
5. Xba1 (with seal)
6. Spe1
7. Pst1
8. 1kb ladder
PCR and Electrophoresis
| Quick Taq Dye Mix | primer-f | primer-r | template | MilliQ | total |
|---|---|---|---|---|---|
| 25 | 1.0 | 1.0 | 0.5 | 22.5 | 50 |
Predenature 94℃,2min
Denature 94℃,30sec
Annealing 59℃,30sec
Extension 68℃,3min
→25cycles
Restriction
| BufferH | tatABCD | EcoR1 | Spe1 | MilliQ | total |
|---|---|---|---|---|---|
| 2 | 5 | 0.2 | 0.2 | 12.6 | 20 |
PCR purification
We eluted the product for 30µL MilliQ
Ligation
| Insert(tatABCD) | Vector(pSB1C3) | Ligation High | total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
4℃, overnight
February 29
Transformation
| tatABCD | competent cell | total |
|---|---|---|
| 1 | 10 | 11 |
Checking Restriction enzyme
| plasmid seems to be 1-18C promoter | Enzyme | Buffer | MilliQ | total |
|---|---|---|---|---|
| 2 | 0.2 | 2 | 15.8 | 20 |
Checking tatABCD
| tatABCD | Hind3 | Buffer | MilliQ | total |
|---|---|---|---|---|
| 5 | 0.2 | 2 | 12.8 | 20 |
PCR
| buffer | dNTPs | MgSO4 | Primer-f | Primer-r | ColE1(6.5ng/µL) / TMAO | MilliQ | KOD Plus Neo | total | |
|---|---|---|---|---|---|---|---|---|---|
| Kil | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16 | 0.5 | 25 |
| TMAO | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16 | 0.5 | 25 |
Predenature 94℃,2min
Denature 98℃,10sec
Annealing 60℃,30sec
Extension 68℃,3min
→30cycles
Electrophoresis
1. 1kb ladder
2. Kil (649bp)
3. TMAO (2720bp)
4. TMAO (Quick Taq)
5. tatABCD (Quick Taq)
6. tatABCD (Hind3)
7. 1kb ladder
March 1
PCR
- TMAO
Template is gDNA and product of colony PCR gel extraction
| Buffer | gNTPs | MgSO4 | Primer-f | Primer-r | KOD Plus Neo | Template gDNA | product of gel extraction | DW | total | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 0.5 | 0 | 16 | 25 |
| 2 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 0 | 2 | 14.5 | 25 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 1.5min
→25cycles
- Kil
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | colE1(6.5ng/µL) | KOD Plus Neo | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16 | 0.5 | 25 |
94℃, 2min
98℃, 10sec
61℃, 30sec
68℃, 1min
→20cycles
Electrophoresis
1. 1kb ladder
2. TMAO1 (gDNA)
3. TMAO2 (product of gel extraction)
4. Kil
PCR
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | Product of Purification | KOD Plus Neo | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 32 | 1 | 50 |
94℃, 2min
98℃, 10sec
61℃, 30sec
68℃, 30sec
20cycles
→Purification 230ng/µL
Restriction
| Kil | EcoR1 | Spe1 | BufferH | MilliQ | total |
|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 2 | 12.6 | 20 |
incubate at 37℃, for 1.5 hours
PCR Purification
Ligation
| Kil | pSB1C3 | Ligation High | total |
|---|---|---|---|
| 5 | 1 | 3 | 9 |
at 4℃, for overnight
March 2
Ligation
| Kil | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 5 | 1 | 3 | 9 |
| tatABCD | pSB1C3 | Ligation | total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
at 16℃ for 1 hour
Transformation
| Kil | Kil(3/1,Ligation) | tatABCD | competet cell | total |
|---|---|---|---|---|
| 1 | 0 | 0 | 10 | 11 |
| 0 | 1 | 0 | 10 | 11 |
| 0 | 0 | 1 | 10 | 11 |
PCR
| Quick Taq | primer-r | primer-f | template | MilliQ | total |
|---|---|---|---|---|---|
| 25 | 1 | 1 | 0.5 | 22.5 | 50 |
Electrophoresis
(27)
Restriction
| pSB3C5-5 | EcoR1 | Pst1 | BufferH | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃, for 2 hour
Electrophoresis
1. 1kb ladder 2µL
2. pSB3C5 5µL + 6×Loading Buffer 1µL
・product of gel extraction(about 2700bp)
-30.9µg/mL
Restriction
GFP1,2,3
| GFP | EcoR1 | Pst1 | Buffer | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
at 37℃, for 2.5 hours
Restriction
| DT | EcoR1 | Xba1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 3 | 0.3 | 16.3 | 30 |
| Constitutive Promoter | Spe1 | Pst1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 15 | 0.2 | 0.2 | 3 | 0.3 | 11.3 | 30 |
at 37℃, 2 hours
- J23117-1:135ng/µL, J23107-1:115ng/µL
- DT3→PCR Purification
- Promoter→Gel Extraction
Checking TMAO
| something seems to be TMAO | Buffer2 | EcoR1 | MilliQ | total |
|---|---|---|---|---|
| 10 | 2 | 0.5 | 7.5 | 20 |
at 37℃, for 1 hour
Electrophoresis
1. 1kb ladder
2. GFP1 that had been cut by restriction enzyme
3. GFP2 that had been cut by restriction enzyme
4. GFP3 that had been cut by restriction enzyme
5. GFP1
6. GFP2
7. GFP3
8. TMAO (control)
9. TMAO (EcoR1)
10. DT (control)
11. DT (EcoR1, Xba1)
12. 1kb ladder
Checking tatABCD
| Quick Taq | primer-f | primer-r | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
Electrophoresis
(28)
March 3
PCR
| template | buffer | dNTPs | MgSO4 | VF | VR | KOD plus | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 5 | 5 | 3 | 1.5 | 1.5 | 1 | 32 | 50 |
| 2 | 2 | 5 | 5 | 3 | 1.5 | 1.5 | 1 | 31 | 50 |
94℃, 2min
98℃, 10sec
50℃, 30sec
68℃, 1min
→30cycles
Miniprep
March 4
Sequence of tatABCD
| Quick Taq | primer-f | promer-r sequence | template | MilliQ | total |
|---|---|---|---|---|---|
| 25 | 1 | 1 | 1 | 23 | 50 |
| Quick Taq | primer-f sequence | primer-r | template | MilliQ | total |
|---|---|---|---|---|---|
| 25 | 1 | 1 | 1 | 23 | 50 |
Colony PCR of TMAO
| buffer | dNTPs | NgSO4 | primer-f | primerr-r | KOD plus | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 4 | 1.5 | 1.5 | 1 | 32 | 50 |
→ethanol precipitation
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 2.5min
→25cycles
Electrophoresis
1. 1kb ladder
2. tatABCD1
3. tatABCD2
4. TMAO
5. 1kb ladder
Restriction
| TMAO | EcoR1 | BufferH | BSA | MilliQ | total |
|---|---|---|---|---|---|
| 10 | 0.2 | 3 | 0.3 | 16.5 | 30 |
| TMAO | Xba1 | Pst1 | BudderM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 3 | 0.3 | 16.3 | 30 |
at 37℃ for 1 hour
Electrophoresis
Transformation
| pSB1C3 | competent cell(made at 2/8) | total |
|---|---|---|
| 5 | 100 | 105 |
March 5
Restriction
| pSB1C3(Xba1, Spe1) | Pst1 | BufferH | BSA | MilliQ | total |
|---|---|---|---|---|---|
| 10 | 0.2 | 2 | 0.2 | 7.6 | 20 |
| pSB1C3(Xba1, Spe1) | EcoR1 | BufferH | BSA | MilliQ | total |
| 10 | 0.2 | 2 | 0.2 | 7.6 | 20 |
at 37℃ for 1 hour
→Then we did ethanol precipitation
Ligation
| Kil(EcoR1, Spe1) | pSB1C3(EcoR1) | Ligation High | total |
|---|---|---|---|
| 5 | 1 | 3 | 9 |
at 16℃ for 1 hour
Transformation
| Kil | competent cell | total |
|---|---|---|
| 1 | 10 | 11 |
We used commercially available competent cells in this time.
PCR
TMAO
| buffer | dNTPs | MgSO4 | Primer-f | Primer-r | Template | KOD plus | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 1 | 32 | 50 |
Electrophoresis
(31)
Restriction
| Lacp | pSB3C5 | EcoR1 | Pst1 | BufferH | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|
| 1 | 20 | 0 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
| 2 | 0 | 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for 1 hour
1→Ethanol precipitation 45.4µg/mL
2→Gel extraction 38.7µg/mL
Ligation
| LacP | pSB3C5 | Ligation High | total |
|---|---|---|---|
| 10 | 2 | 6 | 18 |
at 4℃ for overnight
Transformation
| Lacp+pSB3C5 | competent cell | total |
|---|---|---|
| 1 | 10 | 11 |
on ice for 30 mins.
heat shock at 42℃ for 60secs
on ice for 2 mins.
After we incubate with 200µL of SOC culture for 1 hour, we did plating on LB culture with CP
Restriction
| GFP Plasmid | EcoR1 | Spe1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
at 37℃ for 2 hours
Electrophoresis
1. Ladder 2µL
2. GFP Plasmid (already restricted) 2µL + Loading Dye 2µL + MilliQ 8µL
3. Ladder 2µL
PCR
torA signal and pspA
pspAはコロニーPCR
| Buffer | dNTPs | MgSO4 | Primer-f | Primer-r | template(TMAO) | KOD plus | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 0.5 | 16 | 25 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
electrophoresis
1. 100bp Ladder
2. torA signal (272bp)
3. pspA (969bp)
4. 100bp Ladder
March 6
Restriction
| GFP | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 12 | 0.5 | 0.5 | 3 | 0.5 | 13.5 | 30 |
We did incubate at 37℃ for 1.5hours.
And we did gel extraction on 3/7 and get 40.0μg/mL GFP.
PCR
| buffer | dNTPs | MgSO4 | Primer-f | Primer-r | template | KOD plus neo | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 0.5 | 1 | 32.5 | 50 |
94℃ 2min
98℃ 10sec
60℃ 30sec
68℃ (torA 10sec / pspA 30sec) 25 cycles
We used product of PCR on 3/5 of torA and pspA as template.
Restriction
| Kil | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.3 | 0.3 | 3 | 0.3 | 16.1 | 30 |
| DT | EcoR1 | Xba1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
at 37℃ for 2 hours
→ purification 37.7ng/μL
Ligation
| Kil | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
- Kil : 350fmol
- pSB1C3 : 29fmol
at 16℃ for overnight
March 7
Electrophoresis
1. 1kb ladder 2µL
2. pspA (PCR product)2.5µL + Loading Dye 0.5µL
3. GFP 30µL + Loading Dye 6µL
4. 1kb ladder 2µL
- The GFP was gel extracted on 3/6 and concentrated by Vacuum in 150µg/mL
Ligation
| VectorDNA | GFP | Ligation High Ver.2 | total |
|---|---|---|---|
| 5 | 15 | 10 | 30 |
Restriction
| torA | EcoR1 | Spe1 | bufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.3 | 0.3 | 3 | 0.3 | 16.1 | 30 |
| pspA | EcoR1 | Spe1 | bufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.3 | 0.3 | 3 | 0.3 | 21.1 | 30 |
at 37℃ for 1.5 hours
Purification
torA→31.8ng/µL
pspA→49.3ng/µL
Ligation
| torA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 3 | 3 | 3 | 9 |
| pspA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
at 4℃, for overnight
- torA→31.8ng/µL×3µL=95.4ng=0.529pmol
- pSB1C3→19.4ng/µL×3µL=58.2ng=0.042pmol
- pspA→49.3ng/µL×4µL=197.2ng=0.308pmol
- pSB1C3→19.4ng/µL×2µL=38.8ng=0.029pmol
March 8
Restriction
| pSB4K5 | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 20 | 0.2 | 0.2 | 3 | 0.2 | 6.4 | 30 |
at 37℃ for 1 hour.
→Purification : 36.6ng/µL
Ligation
| Kil | pSB4K5 | Ligation High Ver.2 | total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
at 4℃ for overnight
- Kil→37.7ng/µL×10µL=377ng=879fmol
- pSB4K5→36.6ng/µL×1µL=36.6ng=86fmol
Liquid culture
Lacp + pSB3C5 -1, 2
Transformation
| torA | pspA | competent cell | total |
|---|---|---|---|
| 1 | 0 | 10 | 11 |
| 0 | 1 | 10 | 11 |
We use commercially available competent cells in this time.
March 9
Restriction
| tatABCD | Xba1 | Pst1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 3 | 0.3 | 16.3 | 30 |
at 37℃ for 1hour
→purification : 75.0μg/mL (1.11 260/280 , 0.81 260/230)
Miniprep
lacP + pSB3C5 1 80.5μg/mL (1.78 260/280 , 2.00 260/230)
lacP + pSB3C5 2 107.2μg/mL (1.83 260/280 , 1.90 260/230)
Colony PCR
| Quick Taq | VF | VR | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃ 2min
94℃ 30sec
55℃ 30sec
68℃ 6sec
25cycles
Ligation
| tatABCD | constP J23107 | Ligation High Ver.2 | total |
|---|---|---|---|
| 5 | 1 | 3 | 9 |
tatABCD : 227fmol
constP J23107 : 21fmol
Transformation
| pspA | torA | Kil | competent cell | |
|---|---|---|---|---|
| 1 | 1 | 0 | 0 | 10 |
| 2 | 0 | 1 | 0 | 10 |
| 3 | 0 | 0 | 1 | 10 |
Miniprep
4mL of plusgrow which had been cultured for overnight.
pSB4K5 : 80.5μg/mL
March 10
Screening PCR
| Quick Taq | VF | VR | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
Then we did electrophoresis to confirm.
1.1kb ladder
2.kil (649bp)
3,4,5, pspA (969bp)
6,1kb ladder
1, 100bp ladder
2,3,4, torA signal
Restriction
| LacP-pSB3C5 | Spe1 | Pst1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 2 | 0.2 | 7.4 | 20 |
for 2.5 hours at 37℃
→purification 29.0ng/μL
| torA | Xba1 | Pst1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 2 | 0.2 | 7.4 | 20 |
for 2.5 hours at 37℃
→purification 91.8ng/μL
Ligation
| torA | Lacp-pSB3C5 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
torA : 929fmol
Lacp-pSB3C5 : 284fmol
for overnight at 4℃
March 11
Miniprep
We used 3μL of plus grow that we had cultured for overnight.
torA : 62.8ng/μL
Screening PCR
| Quick Taq | VF | VR | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
Electrophoresis
1. 1kb ladder
2〜11. pspA (969bp)
12. 1kb ladder
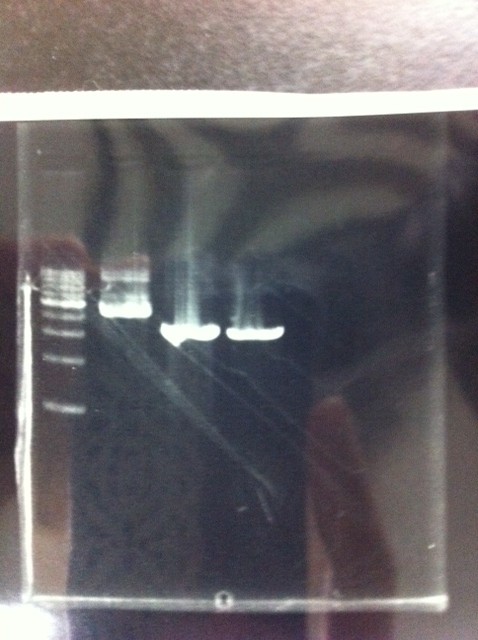
The results were shown as photograph in the right.
It seemed that there were shorter sample than expected sample, so we did electrophoresis with pspA which was product of PCR and pspA which had already cut with EcoR1 and Spe1.
1.1kb ladder
2. pspA (PCR product)
3, pspA (Eco, Spe)
4〜6, pspA (colony PCR)
7,1kb ladder
The results were shown as photograph in the right.
March 12
Transformation
| DT(1ng/μL) | DT(0.1ng/μL) | Kil | lacP-torA | MilliQ | competent cell | total |
|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 20 | 21 |
| 0 | 1 | 0 | 0 | 0 | 20 | 21 |
| 0 | 0 | 5 | 0 | 0 | 50 | 51 |
| 0 | 0 | 0 | 5 | 0 | 50 | 51 |
| 0 | 0 | 0 | 0 | 1 | 20 | 21 |
Restriction
| pspA | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.3 | 15.7 | 30 |
at 37℃ for 4 hours
→ We did purification and got 48.3ng/μL pspA.
Ligation
| pspA | pSB1C3 | MilliQ | Ligation High Ver.2 | total |
|---|---|---|---|---|
| 4 | 2 | 0 | 3 | 9 |
| 2 | 2 | 0 | 2 | 6 |
| 0 | 2 | 2 | 2 | 6 |
March 13
Miniprep
pSB1C3 74.2µg/mL 1.65 (260/280) 1.31 (260/230)
March 14
Restriction
| pSB1C3 | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 20 | 0.2 | 0.2 | 4 | 0.4 | 15.2 | 40 |
We did gel extraction and got 47.2ng/µL pSB1C3 but we did not cut out RFP.
Ligation
| torA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
| MilliQ | pSB1C3 | Ligation High Ver.2 | total |
| 4 | 2 | 3 | 9 |
at 16℃, for 1 hour
- torA : 0.707pmol
- pSB1C3 : 0.068pmol
Liquid culture
We cultured Lacp-pSB3C5 1,2,3,4,5 (CP tolerance)on culture with Amp.
→Only 4 which did not be cultured succeeded.
March 15
Liquid culture
We cultured Lacp-pSB3C5 6,7,8,9,10 on culture with ampicillin.
→6,8,9,10 were succeeded.
Restriction
| GFP | EcoR1 | Spe1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
| DT | EcoR1 | Xba1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
for 2 hours at 37℃.
Miniprep
pspA (pSB1C3) 40.5ng/µL
Ligation
| pspA | DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 5 | 1.5 | 3 | 9.5 |
- pspA : 385fmol
- DT : 36fmol
Transformation
| J23107-tatABCD | DT (0.1ng/µL) | DT (0.01ng/µL) | pspA-DT | competent cells on 3/15 | total |
|---|---|---|---|---|---|
| 2 | 0 | 0 | 0 | 20 | 22 |
| 0 | 2 | 0 | 0 | 20 | 22 |
| 0 | 0 | 2 | 0 | 20 | 22 |
| 0 | 0 | 0 | 2 | 20 | 22 |
Screening PCR
Kil, pspA and torA
| Quick Taq | Primer-r | Primer-f | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
March 16
Miniprep
torA (pSB1C3) 68.8ng/µL
Kil (pSB4K5) 92.7ng/µL
torA was red for some reason. We do not know why.
Colony PCR
| Quick Taq | Primer-r | Primer-f | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 6sec
→25cycles
Electrophoresis
The results were shown as photograph in the right.
Checking Transformation Efficiency
competent cells that were made on March 15.
DNA : 0.02ng → 668 colonies Transformation Efficiency : 3.3×10^7
DNA : 0.2ng → 1739 colonies Transformation Efficiency : 8.7×10^6
Restriction
| pSB1C3 | EcoR1 | Spe1 | BSA | BufferM | BufferH | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 20 | 0.2 | 0.2 | 0.3 | 3 | 0 | 6.3 | 30 |
| 5 | 0.2 | 0 | 0.2 | 0 | 2 | 12.6 | 20 |
at 37℃, for 2 hours.
We did gel extraction for product with EcoR1, Spe1. We got 46.1ng/µL pSB1C3.
| Kil(pSB4K5) | EcoR1 | Pst1 | BSA | BufferH | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
for overnight at 37℃.
We did this to confirm.
| pspA (pSB1C3) | EcoR1 | Pst1 | BSA | BufferH | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
for overnight at 37℃.
We did this to confirm.
Ligation
| torA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
| MilliQ | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
at 16℃, for overnight
- torA→767fmol
- pSB1C3→68fmol
March 17
Miniprep
J23107-tatABCD 72.7ng/µL
pspA-DT 50.5ng/µL
Checking the Insert
| J21037-tatABCD | EcoR1 | Pst1 | BSA | BufferH | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
Success.
| pspA-DT | EcoR1 | Pst1 | BSA | BufferH | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
Failed.
March 19
Restriction
| DT | EcoR1 | Xba1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
We did Gel extraction and got 17.2ng/µL of DT.
Ligation
| Kil | DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 10 | 2 | 6 | 18 |
We did this for an hour at 16℃.
Restriction
| GFP | EcoR1 | Spe1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 0.5 | 3 | 15.5 | 30 |
We did this for 4 hours at 37℃
Ligation
| pspA | DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 5 | 5 | 5 | 15 |
| pspA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 1 | 3 | 8 |
We did these for an hour at 16℃.
- pspA (5µL)→377fmol
- DT→39fmol
- pspA (4µL)→339fmol
- pSB1C3→34fmol
Transformation
pspA-DT, pspA (pSB1C3), torA (pSB1C3) and GFP-DT
March 20
Screaning PCR
| Quick Taq | Primer-R | Primer-F | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
- pspA→○
- pspA-DT→○
- GFP-DT→○
- torA→×
- Kil-DT 6 of 8 sumples→○
| Quick Taq | VR | VF | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
- pspA→○
- pspA-DT→×
Restriction
| torA | EcoR1 | Spe1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.3 | 0.3 | 0.3 | 3 | 16.1 | 30 |
| DT | EcoR1 | Xba1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
We did this for overnight at 37℃. And we did purification.
torA 34.2ng/µL
DT 28.0ng/µL
March 21
Miniprep
GFP-DT-1 : 77.7ng/µL
GFP-DT-2 : 67.6ng/µL
Restriction
| pSB1C3 | EcoR1 | Spe1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
We did this for 3 hours at 37℃, and then we did gel extraction. We got 25.1ng/µL pSB1C3
| GFP-DT | EcoR1 | Pst1 | Xba1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0 | 0.2 | 2 | 12.6 | 20 |
| 10 | 0.2 | 0 | 0.2 | 0.3 | 3 | 16.3 | 30 |
We did this for 3 hours at 37℃, and then we did Purification. We got 30.7ng/µL GFP-DT.
Ligation
| torA | pSB1C3 | pspA | DT | GFP-DT | Ligation High Ver.2 | total | |
|---|---|---|---|---|---|---|---|
| 1 | 4 | 1 | 0 | 0 | 0 | 3 | 8 |
| 2 | 0 | 1 | 7 | 0 | 0 | 4 | 12 |
| 3 | 0 | 0 | 5 | 3 | 0 | 4 | 12 |
| 4 | 3 | 0 | 0 | 0 | 5 | 4 | 12 |
We did this for an hour at16℃.
March 22
PCR
We did PCR to amplify torA_signal that was product of PCR at March 5 with redesigned primers.
| Buffer | dNTPs | MgSO4 | Primer-F | Primer-R | Template | MilliQ | KOD plus neo | total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 0.5 | 32.5 | 1 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 10sec
→30cycles
→Purification 110.7ng/µL
Restriction
| torA | EcoR1 | Spe1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
Ligation
| torA | pSSB1C3 | pspA | DT | GFP-DT | Ligation high | total | |
|---|---|---|---|---|---|---|---|
| 1 | 4 | 3 | 0 | 0 | 0 | 4 | 11 |
| 2 | 3 | 0 | 0 | 0 | 3 | 3 | 9 |
| 3 | 0 | 0 | 5 | 5 | 0 | 5 | 15 |
- torA (4µL)→512fmol
- pSB1C3→54fmol
- torA (3µL)→384fmol
- GFP-DT→36fmol
- pspA→377fmol
- DT→65fmol
March 23
Screening PCR
torA (pSB1C3), torA-GFP-DT and pspA-DT
| Quick Taq | VF | VR | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
E.coli in liquid culture that had pspA(pSB1C3) also expressed RFP.
Restriction
| pspA | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.3 | 0.3 | 3 | 0.3 | 21.1 | 30 |
And then we did ethanol precipitation
Ethanol precipitation
pspA 11.5ng/µL.
Miniprep
Lacp+pSB3C5-8 77.9µg/mL
Lacp+pSB3C5-10 69.0µg/mL
Kil+DT-4 58.0µg/mL
Kil+DT-7 15.2µg/mL
Restriction
| Lacp+pSB3C5-8 | Spe1 | Pst1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
| Kil+DT-4 | Xba1 | Pst1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for 1.5 hours
And then we did Gel extraction.
Gel Extraction
Lacp+pSB3C5-8 40.4µg/mL
Kil+DT-4 26.8µg/mL
March 26
Miniprep
torA(pSB1C3) 18.5[ng/µL]
torA-GFP-DT 20.0[ng/µL]
Restriction
| torA(pSB1C3) | EcoR1 | Pst1 | BufferH | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 2 | 0.2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 2 | 0.2 | 12.6 | 20 |
| torA-GFP-DT | EcoR1 | Xba1 | Pst1 | BufferH | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5 | 0.2 | 0 | 0.2 | 2 | 0 | 0.2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0 | 2 | 0 | 0.2 | 12.6 | 20 |
| 20 | 0 | 0.2 | 0.2 | 0 | 3 | 0.3 | 6.3 | 30 |
We did Gel extraction and then got ??? 28.7[ng/µL]
Ligation
| Lacp (pSB3C5) | torA-GFP-DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
- Lacp : 22fmol
- torA-GFP-DT : 197fmol
| pspA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
| pspA | DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
- pspA : 180fmol
- pSB1C3 : 18fmol
- DT : 16fmol
| LacP(pSB3C5) | Kil-DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
for 2 hours at 16℃
Transformation
| Lacp-Kil-DT | competent cell | total |
|---|---|---|
| 1 | 10 | 11 |
March 27
Miniprep
We retryed miniprep of torA(pSB1C3).
We got torA and its concentration was 39.3[ng/µL].
Transformation
| Name | Well | Sample | Competent Cells | Total | Plate | Colony |
|---|---|---|---|---|---|---|
| BBa_K117004 | 14J(2011 plate2) | 5 | 20 | ? | ? | ? |
We added 100[µL] of culture medium before we started culturing the E.coli.
Screening PCR
| Quick Taq | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
Lacp-torA-GFP-DT 1, 3, 5, 6, 8, 9 was successful.
pspA-DT was failed.
E.coli that had pspA(pSB1C3) did not make any colony.
Lacp-Kil-DT 1,2,3,4,5,6,7,8 was failed.
Liquid culture
Lacp-torA-GFP-DT
July 30
Ligation
| kil+DT(XbaI,PstI) | LacP(SpeI,PstI) | Ligation High | Total |
|---|---|---|---|
| 30 | 6 | 36 | 72 |
July 31
Restriction Enzyme Processing
| torAGFP+DT | 10×M Buffer | BSA | MilliQ | XbaI | PstI | Total |
|---|---|---|---|---|---|---|
| 2.5 | 3.0 | 0.5 | 23.4 | 0.3 | 0.3 | 30.0 |
Liquid Culture
J23113(backbone J61002)
August 1
Restriction Enzyme Processing
| lacP+kil+DT | BSA | 10×H Buffer | EcoRI | PstI | MilliQ | Total |
|---|---|---|---|---|---|---|
| 5.0 | 0.5 | 3.0 | 0.5 | 0.5 | 20.5 | 30.0 |
| lacP middle copy | BSA | 10×H Buffer | EcoRI | PstI | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10.0 | 0.5 | 3.0 | 0.5 | 0.5 | 15.5 | 30.0 |
MIniprep
J23113(backbone J61002)
Liquid culture
J23113(backboneJ61002) Three test tubes of 4 mL LB medium with ampicillin 37℃ overnight
August 2
August 3
Restriction Enzyme processing
| pSB4K5 | BSA | 10×H Buffer | EcoRI | PstI | MilliQ | Total |
|---|---|---|---|---|---|---|
| 8.0 | 0.5 | 3.0 | 0.5 | 0.5 | 17.5 | 30.0 |
| B0034 | BSA | 10×H Buffer | SpeI | PstI | MilliQ | Total |
|---|---|---|---|---|---|---|
| 12.0 | 0.5 | 3.0 | 0.5 | 0.5 | 13.5 | 30.0 |
DNA purification
Ligation
| lacP+kil+DT | pSB4K5 | ligation high | Total |
|---|---|---|---|
| 15 | 15 | 15 | 45 |
Miniprep
- J23113(backbone J61002) 218.0μg/mL
- J23113(backbone J61002) 252.5μg/mL
- J23113(backbone J61002) 201.0μg/mL
- pSB3C5 45.0μg/ml 1.60 260/280 1.80 260/230
- pSB4C5 213.0μg/mL 1.77 260/280 4.40 260/230
August 4
August 5
August 6
August 7
August 8
August 9
August 10
Ligation
| B0034 SpeI PstI | GFP+DT XbaI PstI | ligation high | Total |
|---|---|---|---|
| 2 | 3 | 4 | 9 |
Transformation
- RBS+GFP+DT
- lacP+kil+DT
- RBS(for control)
- To put 2ng DNA in 20μL competent cell and leave it on ice for 30min
- Heat shock for 60s at 42℃
- To leave on ice for 2min
- To spread on ampicillin LB plate
August 11
August 12
August 13
Liquid culture
B0034 3mL ×3
- 37℃ overnight
August 14
Miniprep
- B0034 0μg/mL
- B0034 235.5μg/mL
- B0034 76.5μg/mL
Colony PCR
lacP+kil+DT ×5
| 2×Quick Taq | VF | VF | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
pspA
| 10×Buffer for KOD-Plus-Ver2 | 2mM dNTPs | 25mM MgSO4 | Primer PsPA f-p | Primer PsPA r-s | KOD-Plus- | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
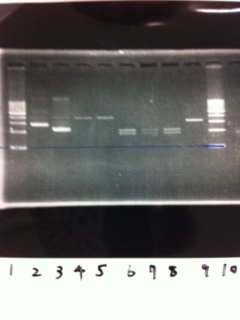
Electrophoresis
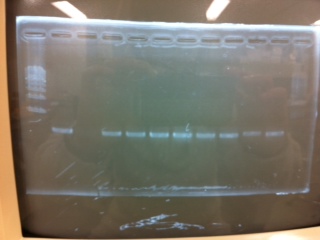
写真貼ってください
positive control(GFP+DT)
negative control(MilliQ)
① ~⑥ colony PCR lacP+kil+DT
次の写真
① 1kb ladder
② Positive control (GFP+DT)
③ Negative control (MilliQ)
④ ~⑨ lacP+kil+DT ①~⑥
⑩~⑫ pspA 10μL ,6×buffer 2μL
⑬ 1kb ladder
August 15
Colony PCR
pspA
| 2×Quick Taq | pspA r-s | pspA f-p | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
56℃, 30sec
68℃, 1min
→35cycles
August 16
August 17
Colony PCR
pspA
| 10×Buffer for KOD-Plus-Ver2 | 2mM dNTPs | 25mM MgSO4 | Primer PsPA f | Primer PsPA r | KOD-Plus- | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 3 | 3 | 1.5 | 1.5 | 1 | 35 | 50 |
| 5 | 3 | 5 | 1.5 | 1.5 | 1 | 33 | 50 |
negative control(MilliQ 50μL)
94℃, 2min
94℃, 15sec
55℃, 30sec
68℃, 1min
→35cycles
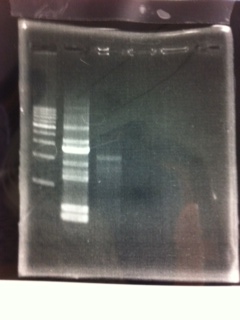
Electrophoresis
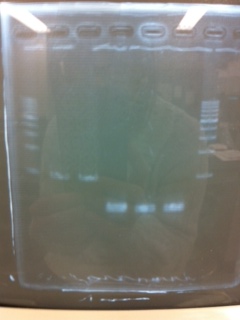
写真貼ってください
① 1kb ladder
②, ③ MgSO4 3μL, pspA
④, ⑤ MgSO4 5μL, pspA
⑥ negative control(MilliQ)
⑧ 1kb ladder
August 18
August 19
August 20
Restriction
| J23113(201.0ng/μL) | Xba1 | Pst1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 0.5 | 14.5 | 30 |
August 21
Colony PCR
pspA
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | Primer PsPA f | Primer PsPA r | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 3 | 5 | 1.5 | 1.5 | 1 | 33 | 50 |
| 5 | 3 | 7 | 1.5 | 1.5 | 1 | 31 | 50 |
| 5 | 3 | 10 | 1.5 | 1.5 | 1 | 28 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
→25cycles
Restriction
| J23113(218ng/μL) | Spe1 | Pst1 | BufferH | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 0.5 | 14.5 | 30 |
| torA-GFP-DT(20ng/μL) | Xba1 | Pst1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.6 | 0.6 | 3 | 0.5 | 20.3 | 30 |
Electrophoresis
写真貼ってください
① 1kb ladder
②, ③ torA-GFP-DT(XbaI,PstI)
④, ⑤ J23113(SpeI,PstI)
Transformation
- lacP-torA-GFP-DT(Backbone pSB3C5)
- BBa_K11704(Backbone pSB1A2)
Colony PCR
- pspA
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | Primer PsPA f | Primer PsPA r | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | Primer PsPA f-p | Primer PsPA r-s | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
- negative control for pspA
We did PCR without a template.
- GFP-DT
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
Transformation
- Lacp-torA-GFP-DT(Backbone pSB3C5)
- J23107-tatABCD
PCR
kil, torA-GFP
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | DNA | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 32 | 1 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
→25cycles
August 22
Restriction
| pSB3C5 | EcoR1 | Pst1 | BufferH | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 13.3 | 0.5 | 0.5 | 3 | 0.5 | 12.2 | 30 |
Gel extraction
pSB3C5(EcoR1, Pst1)
27.2μg/mL
Transformation(the second time)
- Lacp-torA-GFP-DT(Backbone pSB3C5)
- J23107-tatABCD
Electrophoresis
写真お願いします。
See "August 21 -Colony PCR-"
1. 1kb ladder
2. pspA(Primer pspA f&r)
3. negative control for 2
4. GFP-DT
5. pspA(Primer pspA f-p&r-s)
6. pspA(Primer pspA f-p&r-s)
7. negative control for 5&6
PCR
Lacp-GFP-DT(a kit of biological parts)
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | DNA | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 32 | 1 | 50 |
Colony PCR
pspA
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | Primer pspA f | Primer pspA r | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | Primer pspA f-p | Primer pspA r-s | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
→30cycles
Electrophoresis
写真お願いします。
1. 1kb ladder
2. pspA(Primer pspA f&r)
3. pspA(Primer pspA f-p&r-s)
4. negative control for pspA(Primer pspA f&r)
5. negative control for pspA(Primer pspA f-p&r-s)
Transformation
Kil
August 24
Purification of PCR products
pspA(Primer pspA f-p&r-s)
84.1μg/mL
→See "August 22 -Colony PCR-"
 "
"