Team:LMU-Munich/Spore Coat Proteins

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Spore Crust Proteins
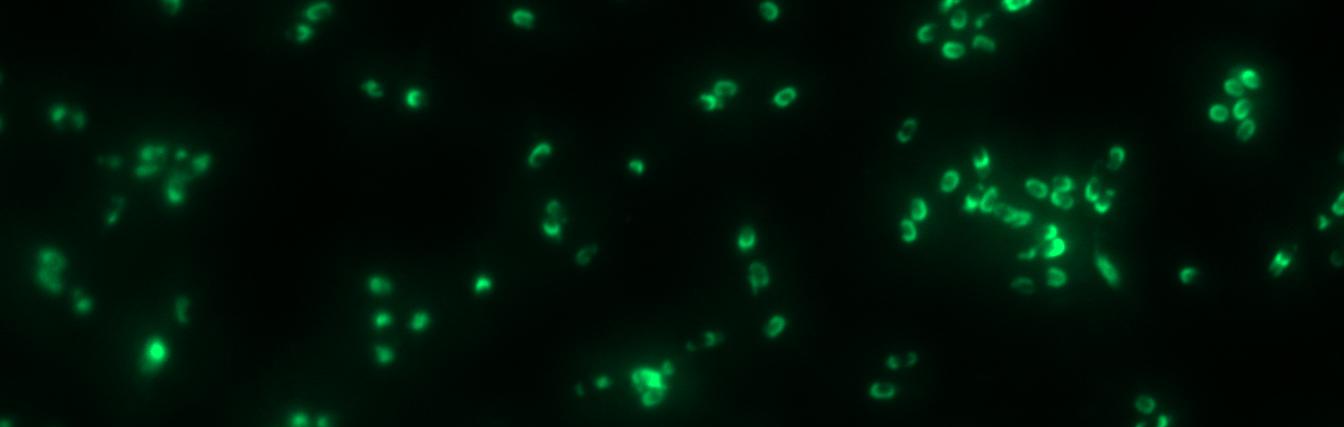
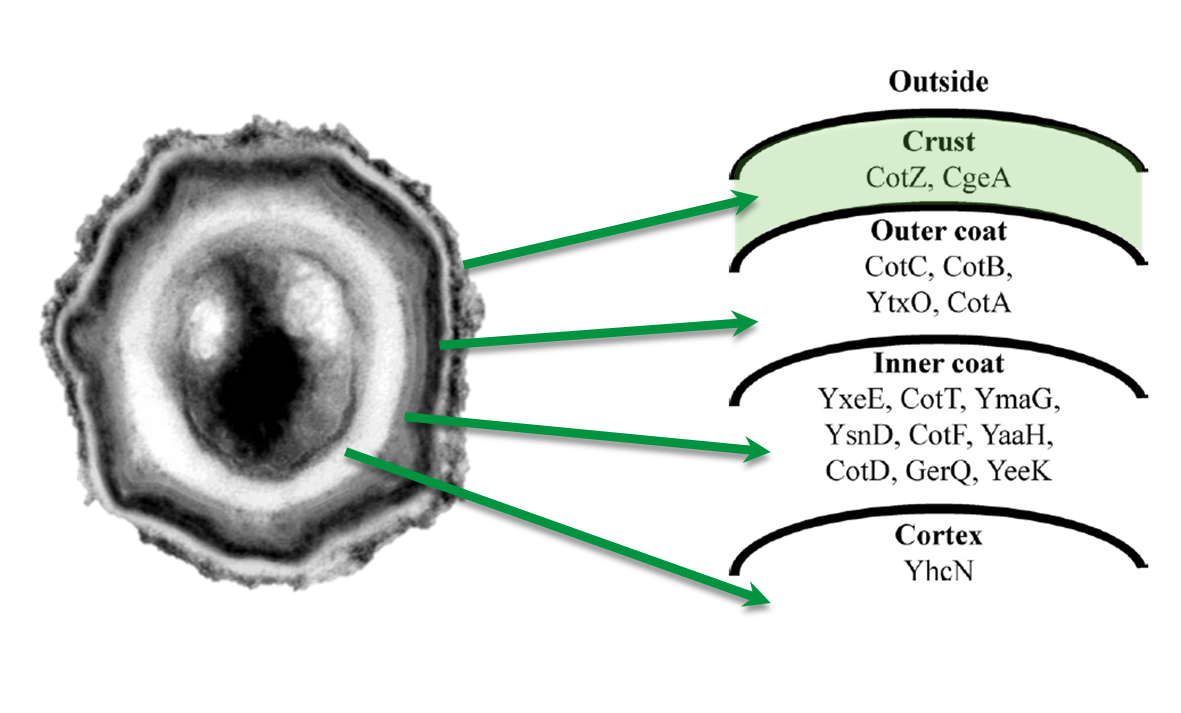
The aim of this project part is to create spores that display fusion proteins on their crust. There are several different proteins forming the spore coat layers of Bacillus subtilis spores. On the outermost layer, the so called spore crust, the CotZ and CgeA proteins are located ([http://www.ncbi.nlm.nih.gov/pubmed?term=imamura%20et%20al.%202011%20spore%20crust Imamura et al., 2011]). This is why we used them to create functional fusion proteins to be expressed on our Sporobeads.
| |
|
The gene cgeA is located in the cgeABCDE cluster and is regulated by its own promoter PcgeA. The cluster cotVWXYZ contains the gene cotZ which is cotranscribed with cotY regulated by the promoter PcotYZ. Another promoter of this cluster PcotV is responsible for the transcription of the other three genes. Those three promoters were evaluated with lux reporter genes to get an impression of their time of activation and their strength (see for more details BacillusBioBrick Box) so they could be used for expression of spore crust fusion proteins.
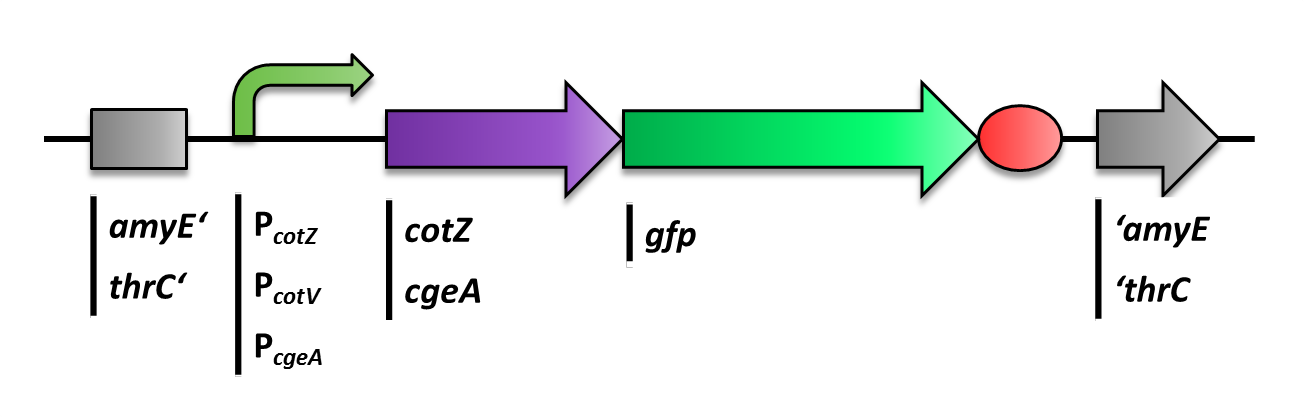
The first step was to fuse gfp to cgeA and cotZ as a proof of principle. This way we would determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation. Therefore we first fused cotZ to its two native promoters, PcotV and PcotYZ, and to PcgeA, which regulates the transcription of cgeA. For cgeA we only used its native promoter PcgeA and the stonger one of the two promoters of the cotVWXYZ cluster, PcotYZ. While gfp was ligated to B0014, a terminator. When these constructs were finished and confirmed by sequencing, we fused them together applying the Freiburg standard to create in frame fusion proteins, flanked by one of the three promoters and the terminator.
| |
|
As we are working with B. subtilis spores, we needed to clone our final constructs into an empty Bacillus vector, so that they could get integrated into the genome of B. subtilis after transformation. Thus we picked the empty vector pSBBS1C from our BacillusBioBrickBox, for the cotZ constructs. This vector integrates into the amyE locus in the B. subtilis genome and therefore we could check the integration of our construct via a starch test. In oder to express both pSBBS4S for cgeA constructs. While the integration of pSBBS1C-cotZ constructs was checked via a starch test, the pSBBS4S-cgeA constructs were tested with threonin-deficient agar plates. The clones with the right integrated constructs have then been chosen for further analysis.
Finally we could start with the important experiment for our GFP-Sporobeads, fluorescence microscopy. Therefore we developed a sporulation protocol, that increases the rates of mature spores in our mutant samples (for details see link). The cells were fixed on agarose-pads and imaged in bright field and blue light.
Project Navigation
| +--+--+--+--+--+--+-- | +--+--+--+--+-- | +--+--+--+--+--+-- | +--+--+--+--+--+-- |

Bacillus BioBrickBOX |

SporeCoat FusionProteins |

Germination STOP |
 "
"