Team:Wellesley HCI/Notebook/KimberlyChang
From 2012.igem.org

Kimberly's Notebook
Contents |
Week of May 28
- May 29: Orientation. Plan. Fill this in
- May 31: I've been looking at the Sifteo documentation and downloading the programs needed to program and run Sifteo applications. These included Siftrunner, Siftdev, Siftulator and MonoDevelop. I also found a basic tutorial on Sean Voisen's blog about getting images to display on the cubes (Media:http://sean.voisen.org/blog/2011/10/up-and-running-with-sifteo-sdk/) By following Sean's tutorial, I was able to get a series of letters to display on the Sfiteo cubes.
Week of June 4
- June 4: Today, our lab visited the Microsoft New England Research and Development Center. There, Edwin and Chris demonstrated Windows 8 running on the Samsung Slate tablet computer. Some of the built-in gestures seemed unintuitive to me. Although users are eventually supposed to become more familiar with the interface, I think that the initial hurdle might be too large for some users. Also because Windows 8 also supports a Desktop environment along with their Metro tiles interface, users will need an external mouse and keyboard in order to interact in the Desktop environment. This seems unwieldy and impractical.
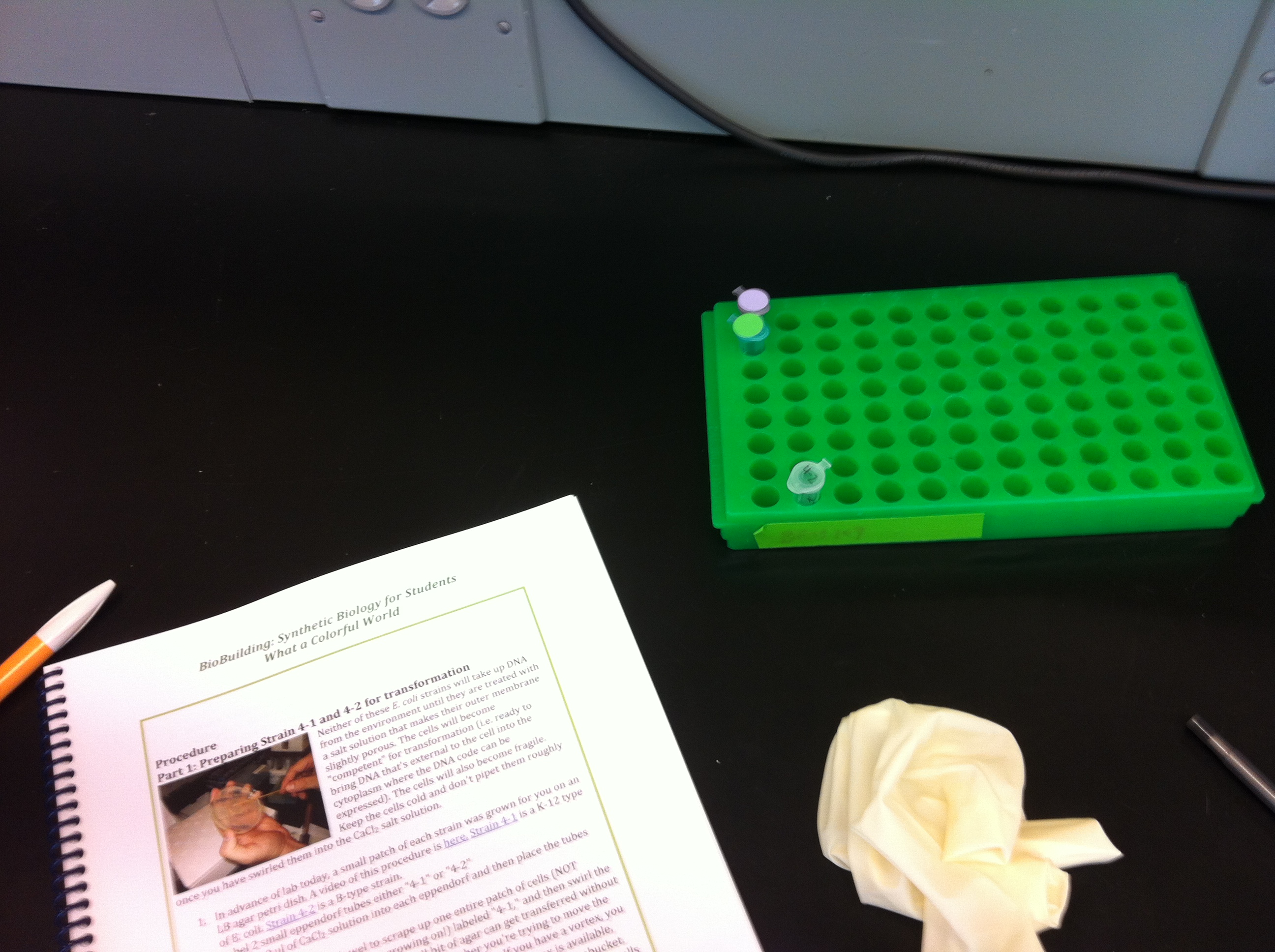
- June 6: The first part of the day was a lecture by Natalie Kuldell explaining some of the basics of synthetic biology. Professor Kuldell showed us a video of the 2006 MIT team presenting their iGEM project. The team's project, Eau d'E coli, involved manipulating E. coli to smell like mint and banana, and the scents would be released at different times during the growth cycle. Because I don't have a strong biology background, there was a lot of terminology I didn’t understand during the presentation. During Professor Kuldell's presentation, the issue of ethics and problems of regulation were also brought up. For instance, people making decisions about synthetic biology are generally uninformed. Also, because the field is so new, there is no government regulation for synthetic biology research. After Professor Kuldell's talk, we had a safety briefing about general lab safety. At MIT, the labs are pressurized so that air always rushes in instead of out. There are also different safety levels in the lab, which are designed to deal with different types of organisms – MIT has 1, 2 and 2.5 levels. The later part of the day was spent completing 2 experiments in the lab. We split into pairs, with one inexperienced person paired up with someone with lab experience. I worked with Kara and learned how to use an automatic pipette and other lab equipment. It was a little overwhelming at first because everything was new to me. Eventually I settled into a rhythm. There was so much specialized equipment used during the lab (such as pipettes, pipette tips, cubettes, eppendorf flasks.) Also, the units of measurement (mics/uL) used in the lab were unfamiliar to me. The most stressful part during the experiments was the part where we had to stop the some chemical reactions every 15 minutes. Overall, the process was very tedious and mechanical.
- June 7: Today I looked at programming for the Surface. I followed a tutorial for creating a simple application called HelloScatterView in Microsoft Visual Studio. The application displays a series of images on the Surface that the user can then manipulate.
Week of June 11
- June 12: BU day!
- June 13: Today we set up a brainstorming image map in an unused classroom upstairs.
- June 15: Today we decide to include flipping the cubes to represent cold shock and shaking the cubes to represent the vortexing and integrating the plasmid into the E. coli. I also attended a Bioethics talk where I learned about the plagiarism problems plaguing the research field.
Week of June 18
- June 18: Today we finalized our paper prototype. We roughed out the basic interactions with the cubes and the Surface using paper cutout Sifteo “screens” and a “storyboard” using posterboard and Post-it notes. We then gave a brief rundown of the installation to Consuelo. While we were demonstrating the project, Linda came up with a good way to deal with the menu cube, as we had been struggling with a good way to implement a menu while keeping the number of specialized cubes to a minimum. Linda’s suggestion was that we utilize the four different sides of the cubes as menu selectors, and depending on which of the sides another cube was placed next to, the menu selection would change.
- June 19: Wendy and I met with Orit to go over our paper prototype. Before meeting with her, we’d come up with different ideas for the installation to connect more directly with synthetic biology. One of these ideas was to create representations of ice buckets and petri dishes that users would then physically “place” the cubes into to cold shock or incubate the bacteria. We thought about using another Surface and possibly even the Microsoft tablet. However, after talking with Orit, we decided that these additional features might be cumbersome. Orit stressed that we needed to grab the attention of our users in about five seconds, so streamlining the interaction was really key. This meant that we needed to simplify the characteristics we gave our creatures and that we needed to get rid of our menu. We are most likely going to be sticking strictly to color as the trait that we are going to modify in order to simplify the interaction further. We are additionally going to move away from a “cute” aesthetic towards something more mature-looking in order to appeal to the more adult audience at iGEM. We discussed testing the prototype installation at Wellesley and also potentially at the Discovery Museum. For the next stage of the project, we need to think about affordance – basically customizing the cubes to distinguish them – and the aesthetics of the installation. My next task is to work on how the E. coli will look and start creating a short animation of the installation by Friday.
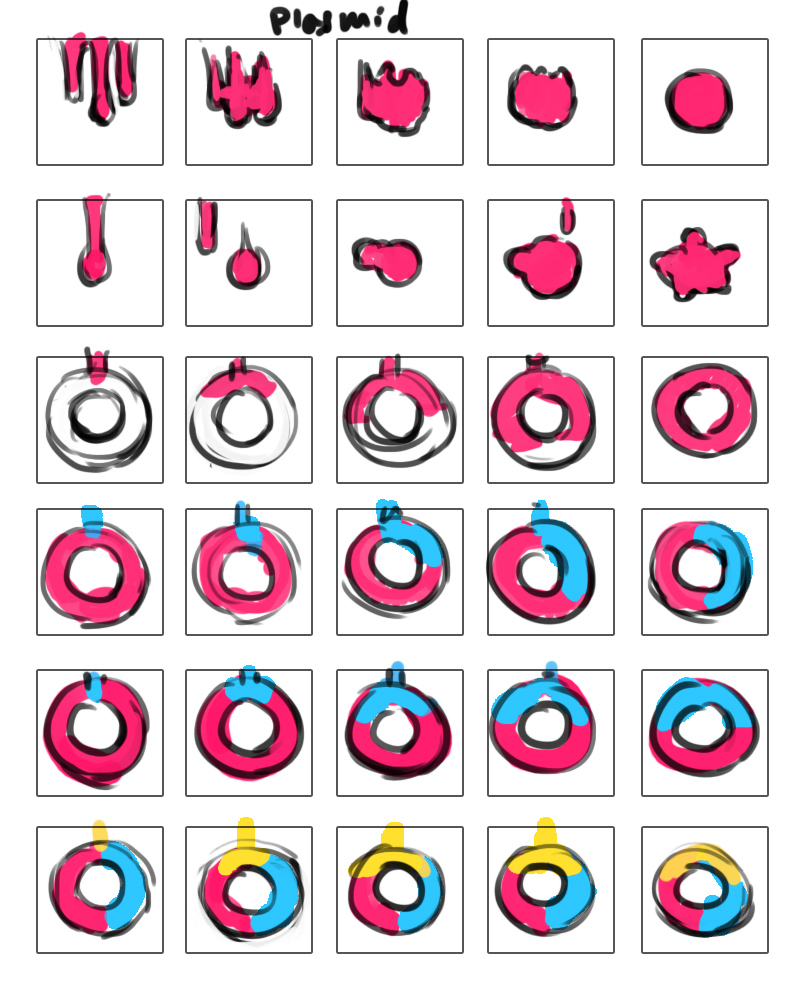
- June 21: I finished creating mockup screens of most of the screens we will need. One of the problems we’re struggling with is how we will incorporate a “tutorial” aspect to the installation to help people figure out how to use the cubes. I came up with several different options for how we can tackle that issue. I can also created mockups of the Surface. I developed three options for what the trails of the E. coli will look like. Meanwhile, Wendy had been thinking about what kind of housing the cubes would have, as we plan to “hack” the cubes by taking them apart and fitting them with custom casings that resemble petri dishes, beakers and the like to simulate a lab environment. We talked with Orit, who seemed very excited by the progress we’ve made. She suggested that we make the plasmid circular to better resemble what actual plasmids look like. She also thought that if we decided to keep the “malfunctioning” creature, we should make it do something interesting. That way, people wouldn’t be as annoyed when their experiment failed.
- June 22: Today was a frustrating day of trying to figure out how to display images. The goal was to get a gif animation to play on the cubes, but after I encountered a lot of difficulty getting that to work, I tried focusing on just getting one image to display. I followed the tutorial on Sean Voisen’s page, but when I tried to substitute my own image, the program stopped working. We tried making new projects, altering the source images, pretty much everything we could think of, but nothing worked.
Week of June 25
- June 25: I decided to take a break from getting an image to display and switched to working on the code that would let users select from a series of colors. I looked at the demo files included from the Sifteo SDK and borrowed code from the Slideshow app. The Slideshow allowed users to cycle through a series of images by clicking, but it also included functionality to affect the images on-screen by shaking and connecting cubes to other cubes. I took the events other than the clicking, but was frustrated to discover that the code wouldn’t run, even when I had copy and pasted the exact same code from the demo file.
- June 26: I looked at the code I was working on from yesterday, and discovered that the demo file had a “bootstrap” file that include the run command. I created a bootstrap file for my project and was happy to discover that the code now ran smoothly. I then created a new file for the gif animation and took the code for cycling through the frames from the color program and was able to create a moving gif animation on the cubes. I also was able to get a blue square to appear on the screen. This is a placeholder for the ice cubes that we’ll be adding for the cold shock. When Wendy and I met with Orit, Orit discussed using the cubes to interact with the Surface. We had wanted to have the user to simply place the cube down on the surface to deploy the E. coli, but since we needed the cubes to recognize some event, we decided that it might be better for the user to place the cubes face down on the Surface to deploy instead.
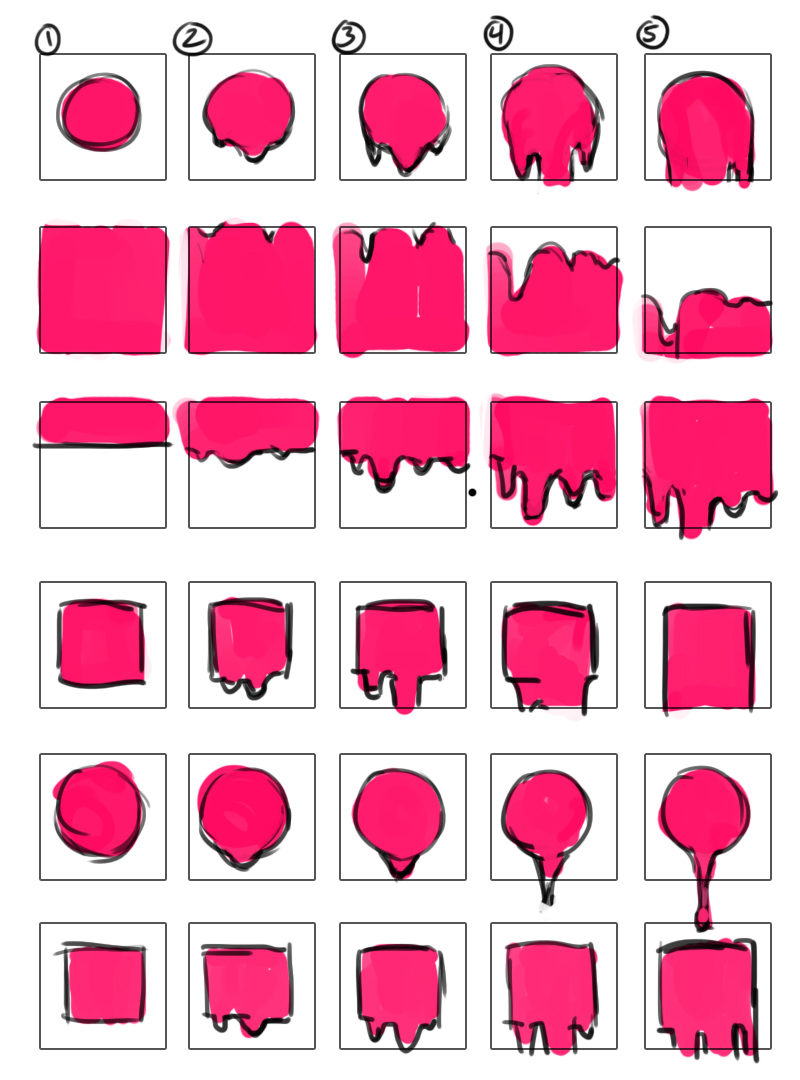
My next task that Orit assigned to me was to get the animation for the dripping paint.
- June 27: I created storyboards for what the dripping animation would look like and how the plasmid cubes would be affected. I didn’t get much done in the way of coding, however.
- June 29: We attended a lecture by Walter Lewin at MIT with members of the BU and MIT iGEM teams. Afterwards, we toured MIT’s iGEM and synthetic biology labs.
Week of July 2
- July 5: Today, Consuelo told me that Nicole will be helping me with the Sifteo cube programming. I sent Nicole the code that I had so far and walked through what I had and what I still wanted to do. Nicole was able to get the tilting interaction working so that when the cubes have been neighbored and the color cube tilts, the dripping animation on both the color cube and the plasmid cube play. I created images for the different colored drips, but we haven’t yet implemented thte code for changing the drip images when the color changes.
- July 6: Nicole was able to get the color cube and plasmid cube drip images to change color to match the color on the color selection cube. I worked on rough animation mockups of the dripping animation and the E. coli animation. I was concerned about using a pre-existing image that had been made by other people for the E. coli image, so I decided to draw my own version. Orit looked over our progress so far and seemed to like what we had. She liked the E. coli animation that I’d come up with and suggested that I make the E. coli resemble the iGEM logo. She also said that ideally we should have a working demo of the app by late next week, so our next task is to incorporate the E. coli cube and its interactions with the other cubes. My goal is to finish the animations for the dripping and the E. coli by Monday.
Week of July 9
- July 9: Over the weekend, I had finished the rough frames of the animation, and today I worked on roughs of the different colors for the Ecoli and the drip images. Nicole and I were having problems getting the images to display on the cubes, however. I decided to at least have the final images complete, so I worked on the final Ecoli images in the evening.
- July 10: Nicole was able to get the images to display, and I finished up the Ecoli images. We ran into a problem with the drip images because they were being drawn over each other instead of refreshing the screen each time. Therefore, I made new drip images that also included the color and plasmid images so that the whole screen would essentially be redrawn every time the drip images were called. We ran into problems with the frame rate and the Ecoli animation moving too quickly on the cubes. In an attempt to solve that, I duplicated each frame of the Ecoli animations 5 times. However, the images are still displaying a little too quickly and we’re not sure how to completely resolve that problem. Another issue we’re having is that for some reason, the Ecoli images are called out of order, so the animation frames are out of order as well. We’re not sure why that’s happening. We met with Orit and explained these issues to her. She didn’t have any major problems with what we have right now, though she did tell Nicole that because she wants to overhaul the code and fix the tilt animation, she should overhaul the code first, otherwise she would end up doing the work twice.
- July 11: Today I worked on more graphics for the Sifteo cubes. I finished up the tilt, shake and ice cube graphics and started on some of the alerts. Nicole, meanwhile, was able to get the Cube images in order so that the animations would play correctly.
- July 12: I realized that the animations for the alerts we need will depend on what the final form of the cubes looks like. Since we are planning on housing the cubes in custom casing, I realized that if I started working on the animations now, without knowing what the cubes’ final forms would be, I could end up with a lot of extra work later to change the animations. I talked to Wendy about this and she suggested that I start working on the redesign of the iGEM wiki. I met with Consuelo, and she went through the current iGEM wiki, saying that the 2012 page should be distinguished from the 2011 one, so that people would know that Wellesley was no longer in a team with BU this year. She wanted the font and color scheme changed. If the site was going to be completely overhauled, Consuelo said that she wanted the site to be all on one page, and that we would use JQuery to skip to different sections of the page. She then pointed me to the site onepagelove.com, a showcase of different one-page websites. I spent a while looking at some of the pages on the site and came up with a few concepts of what the redesign could look like. Consuelo late said that she’d like me to come in tomorrow with a rough design of the website.
- July 13: I finished up some rough designs for the website and continued researching JQuery. I did some tutorials on W3School and made some simple code that toggled and hid a paragraph of text when you click on an image.
Week of July 16
- July 16: Today I talked to Consuelo about the designs for our iGEM website. She told me that I didn’t have to worry about the coding so much because that would be a lab effort, but she added that I should work on the color scheme and the any text that’s needed for the final design. We also met with Orit, who told me that for the demo on Thursday, I should formulate some kind of lesson plan for our testers. On Thursday, 6 chemistry class students will be meeting with us to test our applications. These testers will be our pilot study so we don’t need to worry about surveying them. Nicole and I also worked on our Powerpoint for our presentation on Friday.
- July 17: Today, I finished the Powerpoint for our presentation on Friday. I also finished making a rough draft of the lesson plan for our demo on Thursday. Things I still need to add to the demo are: information on how synthetic biology is actually carried out and more information about HCI and computer science.
- July 18: Today we demoed our applications to counselors from International high schools. We gave them an introduction to the lab and showed off the MoClo Planner and the SynFluo Sifteo interactions. Later, the lab went on a trip to Horseneck beach as a CS outing with other computer science research teams.
- July 19: Today 8 high school students came in to demo the SynFluo project. I started out by showing them the Powerpoint presentation, which introduced them to synthetic biology and to our project. Then the students split into two groups. Nicole demoed our project to one group and Consuelo showed the other group the Beast and the vertical Surface. We ran into difficulties with the project and Surface, so we also ended up showing the students more components of the Beast and the vertical Surface. The students seemed very interested in the project and found putting the E. coli into the Surface especially interesting. Some things that I learned from this demo were: the students had difficulty connecting the concepts of synthetic biology with the Sifteo project; the students had had little previous exposure to biology; the students were interested in the idea of touch interfaces and could explore and learn how things work on their own; the students were very cautious with SynFluo because we told them how to use it. These are some things to keep in mind for later. For my part, I will tailor the presentation to fit students with minimal biology knowledge, which should also help enforce the synthetic biology component of our project. In the evening, the whole lab went on a group trip to watch The Dark Knight.
Week of July 23
- July 23: I worked on rehauling the Powepoint presentation for our demoes. I got rid of a lot of the history behind synthetic biology and iGEM and instead focused on trying to make the concepts of synthetic biology easily understandable. I also added a section where I went over basic biology with students, by introducing a bacterial cell and going over important parts like the DNA and the plasmid. We have also decided to changed the name of our project to SynFlo, instead of SynFluo.
- July 24: Today, we demoed SynFlo with two groups of students. I went through the Powerpoints with both groups and Nicole demonstrated the installation. We unfortunately did not get video footage for the first few students that demoed but we got either video or audio footage from the rest of the students. I tried to reinforce concepts of synthetic biology with the students while they were demoing by asking them questions. The students seemed to find the Sifteo/Surface interaction the most compelling. Some of the students were confused as to why they needed to use the plasmid cube to transfer color but were able to learn the “protocol” fairly easily. The students seemed to enjoy interacting with the E. coli once they were on the surface and liked when they changed color. In the evening, I worked on looking up abstracts from the ACM Digital Library to use as references for our own paper for ITS. I focused on projects that were more “artistic” rather than purely functional. I also started writing the abstract for our SynFlo paper.
- July 25: I worked out roughing out the structure of our paper and sketched out a brief introduction. Later Orit went over what we had written and made suggestions about what we should change. She told me that I should work on the abstract last and instead focus on the introduction for now. We are essentially focusing on our paper for ITS for now rather than the poster for the Science Center poster session. Later, I worked on finishing the animations for the traveling E. coli.
- July 26: Today, I finished a rough draft of the introduction and uploaded the new E. coli images. I also worked on editing the introduction. In the afternoon, I went to a graduate school panel where Wellesley alumnae talked about their experiences in grad school.
- July 27: Wendy and I sat down with Orit, where she went over our drafts for the ITS paper.
Week of July 30
- July 30: I worked on writing the Science Center abstracts today. Orit came in twice to look over what I had written and offered suggestions for improvements. I finished up the abstract and submitted it to Wellesley’s website.
- July 31: I finished the designs for the tangibles for the SynFlo contaminants and started working more on the ITS paper. I finished rough drafts of the Related Works, Future Works, User Studies and References sections. Nicole and I took photos of the Surface and the cubes for use in our poster for the Science Center Poster Session. In the evening I found out that the designs for the tangibles that I’d made would not work for the laser cutter. I needed to redraw the designs in Illustrator and export the file as a pdf document. Wendy and I also took more photos of SynFlo using her camera because a lot of the photos Nicole and I took previously looked blurry. I took screenshots from the Upward Bound demo video footage and sent them to Wendy to use for the poster.
- August 1: I got the tangibles printed on the laser cutter. Each different type of tangible was printed on a color of plastic to match with the cutout shapes on the color selection cube. I took photos of the tangibles to use for the poster. I finished putting together what we have for the ITS paper into the ITS format.
 "
"