Team:Cambridge/Project
From 2012.igem.org


Contents |
Overall project
Abstract
Previous iGEM teams have characterised an impressive array of inducible promoters, along with other elements of biosensing circuitry. But, to date, the output from each is not consistent and, in spite of the unifying biobrick standards used, do not necessarily couple together to make integrated test kits. The Cambridge iGEM 2012 team aim to take the true meaning of biobricks to heart, by creating an open and applied biosensor standard available for use by all subsequent teams, as well as, potentially, by industry and researchers in the field.
The biosensor aims to be modular in design, allowing the kits to be tailored to an individual's requirements, and to use light as an output to allow computer interfacing. We aim to use two luciferases, one to give a read-out of the input, and the other to act as a standard to allow fluctuations in colony size to be taken into account. Furthermore, we shall be using B. subtilis as our chassis, with the view to making the most of the spore forming capacity of bacteria to send out desiccated kits with long shelf lives.
Finally, we also plan to create a cheap electronic device together with a mechanical chassis, which would be able to automatically read the information provided by the luciferases (light intenisty and wavelength) and convert them into calibrated digital information which could then be analysed and manipulated computationally.
- Update - We have found a candidate for a new biobrick to be submitted to the registry of parts. Baker et al, in their paper entitled 'Widespread Genetic Switches and Toxicity Resistance Proteins for Fluoride', have identified a fluoride sensitive riboswitch which we, the Cambridge iGEM 2012 team, feel would be an excellent means to test, as proof of concept, our biosensor design.
- Update2 - We have found another new sensor candidate, the Mg2+ riboswitch from Dann III et al's paper 'Structure and Mechanism of a Metal-Sensing Regulatory RNA'.
Implementation
Project Details
As stated above, the main aim of the project was to develop a bio-sensing standard to promote a platform for the development of novel biosensors that may work in a wealth of different ways but that can all be characterised and coupled to an output that is predictable, reliable and most importantly meaningful.
Standardised Outputs
The main idea driving our project is that every biosensor should be coupled to a standard output with its own response curves, to suit the customer's needs. We decided that decoupling the culture/analyte solution from the detection system (e.g. an electronic one) would be a good idea as otherwise the behaviour of the electrode under different conditions might affect the results. Therefore we chose to use light as the signal transducer. This left us with a choice between biofluorescence and bioluminescence.
Whilst fluorescent proteins have been characterised in far greater detail than luciferases, part of the broader aim of the project is for the kit to be as affordable as possible. And given that the equipment to detect the emission spectra of luciferases is cheaper, we decided that a quantitative measurement of bioluminescence was a better option.
One of the greatest problems we seek to overcome in this project is that of consistent readouts. As is always the case with biology, predictability in our biosensing equipment was going to be an issue. To normalise for cell density productivity, it was decided that a ratiometric output would be absolutely necessary if the output from our biosensors was to be meaningful. Drawing on work done by James Brown and the Haseloff lab into reliable, predictable and quantitative ratiometric measurements using fluorescent proteins, we decided to use these principles as the basis of our work with luciferase. We also decided that, as a side experiment and proof of concept, we would attempt to achieve meaningful ratiometric outputs with fluorescent proteins that could be measured with an (all too expensive!) plate reader.
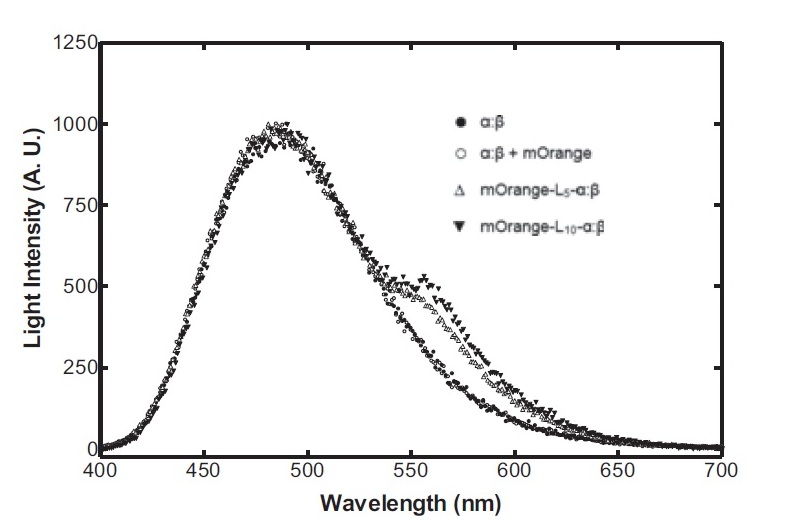
On the luciferase side of things, after a fairly deep trawl through the literature, an OFP-luciferase fusion was found where the emission spectra appeared sufficiently distinguishable from that of the normal bacterial luciferase (a fairly distinctive blue) that it could be measured using simple photo-resistors and coloured filter gels. The emission spectra of the OFP/luciferase fusion is shown below:
The construct that we hope to make is shown below:
Biosensors
As the main crux of this project is a standardised output, we aimed to develop several biosensors, employing different mechanisms, to prove the extended functionality of the final product. To date, most of the biosensors in the registry use an inducible promoter to control expression of their reporter protein. We have used some of these as a proof of the compatability of our kit with a theoretical customers' sensors. However, we also explored another mechanism of biosensing in the form of riboswitches. These have the potential to be the detectors of the future providing a more standard way of designing input circuits and hopefully a faster sensing method as the transciption has already occurred (unlike inducible promoters).
Magnesium riboswitch
Magnesium is essential for life, being a vital component of many enzymatic reactions. Of particular interest for synthetic biology is its role in the action of DNA polymerase enzymes such as Taq. and Phusion. However, no teams have really characterized a sensor that can be used to measure its concentration in solution. Such a biological sensor exists in the form of the bacillus Mg2+ riboswitch. As shown in the diagram, we attempted to isolate this component and submit it as a biobrick, characterizing its function by inserting it into a derepressor construct.
The riboswitch acts as a transcriptional attenuator when Mg2+ is bound, causing disengagement of the RNA polymerase before it can access downstream ORFs. Consequently, these proteins are not expressed. The system that we used inserted this riboswitch just upstream of the LacI repressor in plasmid pJS130. The lac operator that LacI acted on was upstream of sfGFP, consequently expression of LacI blocked transcription of GFP.
To characterize this construct, we will use a 96-well plate reader to assay the effects of the different concentrations of Mg2+ and IPTG on the levels of GFP. We would expect the presence of either to allow the expression of GFP, however because transcriptional attenuation by the riboswitch occurs before expression of the repressor protein, it may be expected that Mg2+ will somehow demonstrate a dominant effect.
Fluoride riboswitch
We also plan on implementing and characterising a Fluoride riboswitch. This, unlike the Magnesium construct, is a positive regulator. The riboswitch, originally isolated from Bacillus cereus, serves as a transcriptional attenuator in the abscence of fluoride. In the presence of fluoride its conformation changes and the repression is lifted. In B. cereus this serves to permit translation of a fluoride efflux pump, which allows the bacteria to cope with the, potentially toxic, elevated fluoride levels in which it finds itself.
Instrumentation
We have constructed a mechanical rotary device that is turned by an arduino-controlled motor to 'sense' from 6 different cuvettes that can be placed in the device and then left for automated detection. The arduino is also connected to two light sensors, one supplied with a blue and the other with an orange filter. The ratio of the light intensity at blue and orange frequencies can be measured to determine the strength of output signal from the bacillus.
The hardware is coupled to a graphical user interface (GUI) that was designed using wxpython. Python is particularly useful as the communication with the arduino microcontroller is done using serial programming, for which python has standard libraries. However, before any communication takes place between the user and the device, the arduino is loaded to perform the basic functions which are written in C++. The arduino and python were chosen for the ease of use and open platform. Also, the arduino is cheap and python is free!
Sporulation and Germination
Another aspect to this project is the longevity of the product. Strains of bacteria expressing various receptors would be generated and stored as dormant spores. This allows the individual tubes of bacterial 'sensors' to sit in the user's cupboard until needed. When the user requires a specific sensor, the appropriate strain is selected and the bacteria can be germinated by following a simple protocol. They can then be placed into the arduino device, the sample loaded and the concentration profile measured.
Part of the main reason for choosing Bacillus subtilis as our chassis was because of it's abiltiy to form dormant spores. As part of our project, we aim to determine a protocol for inducing sporulation in Bacillus, and determine a very simple protocol for germinating the spores so they can be used as part of the system.
It is essential for the germination procedure to be as straight forward as possible, requiring minimal equipment and expertise, so that it could in theory be performed in the field, in a situation where the biosensor might be used.
Results
 "
"