Team:Amsterdam/extra/software
From 2012.igem.org




Software
Contents |
The complexity of biology calls for the aid of computers in a synthetic biology design process. Over the summer we have created a range of useful applications to make the life of the synthetic biologist easier.
Local MediaWiki Editor 0.1
Also tired of using the MediaWiki standard interface for editing? Check out the script we made to make a local copy of the MediaWiki directory structure! It contains features such as uploading/downloading newer files and it works with any editor. Use at your own risk though! since its all automated a settup mistake can result in loss or pollution of data on your wiki.
You can check it out here at git-hub
DNA Template Compiler
Creating custom sequences of DNA is a none trivial task. It requires checks for example RFC-10 forbidden sites, E. coli native restiction and GC contents optimization. This is why we create a script compiles a specially formatted DNA template file into a custom sequences that has been checked and optimized for these parameters.
Click here to check the DNA Template Compiler out at github!
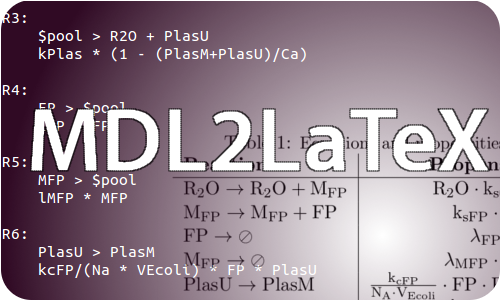
MDL2LaTeX
[http://www.stompy.sourceforge.net StochPy] is a versatile stochastic simulation package implemented in Python for the simulation of biochemical networks. Its inherits the easy to learn model description language [http://www.stompy.sourceforge.net/html/inputfile_doc.html MDL] from its parent package [http://pysces.sourceforge.net/PySCeS PySCeS], which is a modelling environment with deterministic capabilities. By defining models in MDL and executing them through high-level commands in the Python or IPython shell, an easy-going experience is presented to the modeller in which one (almost) doesn’t need to worry about implementation details.
MDL2LaTeX is another step towards making a modeller’s life as carefree as possible. Exactly as its name implies: it converts MDL files to clean LaTeX representations, so the models can be easily integrated into reports. No more time wasted on tedious equation copying into LaTeX!
Installation
MLD2LaTeX has been tested in both Linux and Unix (Mac OS). For easy installation, first install [http://www.github.com git].
Then run:
git clone https://github.com/slagtermaarten/MDL2Latex.git
If you can’t access git, simply go to this MDL2LaTeX github page and download and extract the .zip.
Usage
Edit the
Makefile
inside of this folder and change the pscfile variable to filepath of the the .psc (MDL model) file you want to convert. Save and run
make
 "
"