Membrane Accelerator
Background
As there is no compartment in prokaryotic cells, enzymes involved in a biochemical pathway diffuse all over the cytoplasm. Intermediates produced from one enzyme cannot be passed efficiently to the next due to spatial obstacles.
Rationally designed assemblies of enzymes could help substrates flow by decreasing distance that intermediates have to travel between enzymes, and thus increase yields of sequential biological reactions. If we attach those enzymes to engineered membrane proteins which constitutively assemble together, the reactions are going to proceed much faster.
Construction
As demonstrated before, we built our device on E.coli inner membrane protein Lgt. SRP-dependent signaling sequence of DsbA is fused to N-terminus to localize fusion proteins to membrane.
To rationally assemble and arrange enzymes onto inner membrane of E.coli, we incorporate three protein domains that can interact with their cognate ligands. Protein-protein interaction domains and ligands from metazoan cells (mouse SH3 and PDZ domains and rat GBD domain) were recruited to assemble enzymes on membrane. Each domain could assemble with its cognate ligand.
File:12SJTU 2PDZ.gif
File:12SJTU GBD.gif
File:12SJTU SH3.gif
|_Fig.1 : PDZ domain & ligand __|_Fig.2 : GBD domain & ligand __|___Fig.3 : SH3 domain & ligand__|
|____ from PDB 2PDZ ___________|_______ from PDB 1T84_________|________ from PDB 1CKB_________|
Fluorescence Complementation Assay
To prove whether our membrane protein equipped with those interacting protein could assemble with each other, we conducted fluorescence complementation assay.
EGFP was split into two halves, named 1EGFP and 2EGFP respectively. If there is interaction between two proteins which were fused with 1EGFP and 2EGFP, it is expected that fluorescence should be observed. Otherwise, no fluorescence could be observed.
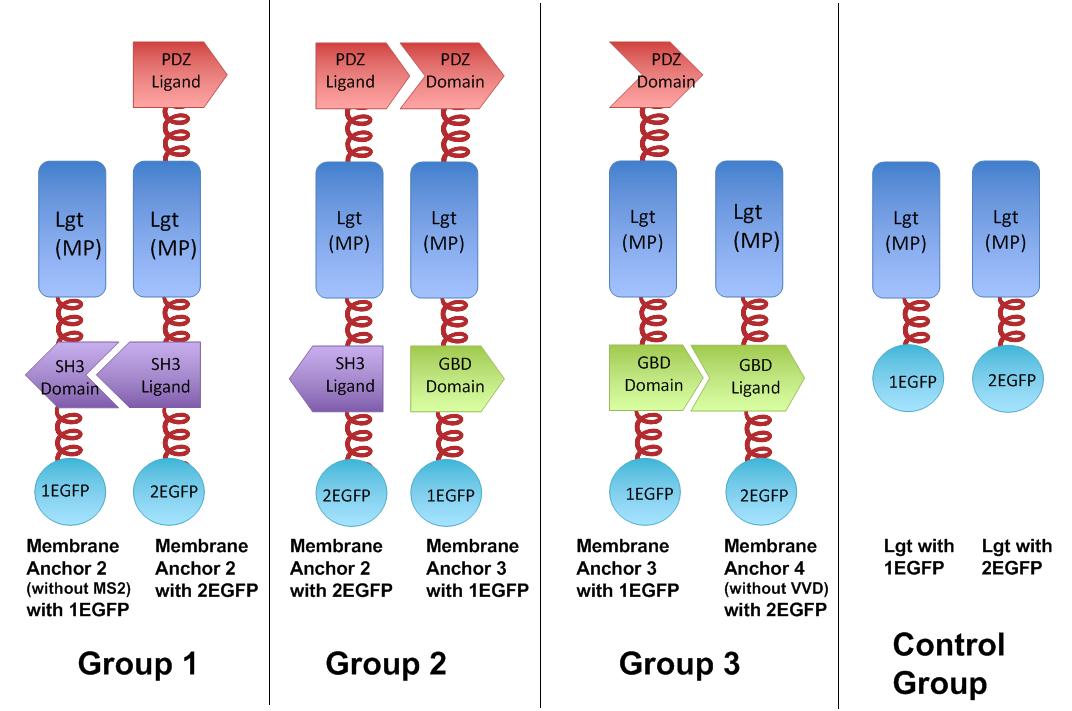 Fig.4 :Group 1, 2, 3 are designed to test the interaction of SH3domain & ligand, PDZdomain & ligand and GBDdomain & ligand respectively. Proteins within each group were expressed in E.coli. If there exists interaction within membrane proteins of each group, we expected to observe fluorescence on E.coli membrane. Control group is supposed to show much weaker fluorescence Proteins within each group are coexpressed in E.coli. L-arabinose is not added into culture media until OD value of bacteria culture reaches 0.6. Bacteria are induced at L-Arabinose concentration of 0.2% for 6 hours. Bacteria samples are smeared onto glass slide and observed under laser confocal microscope.
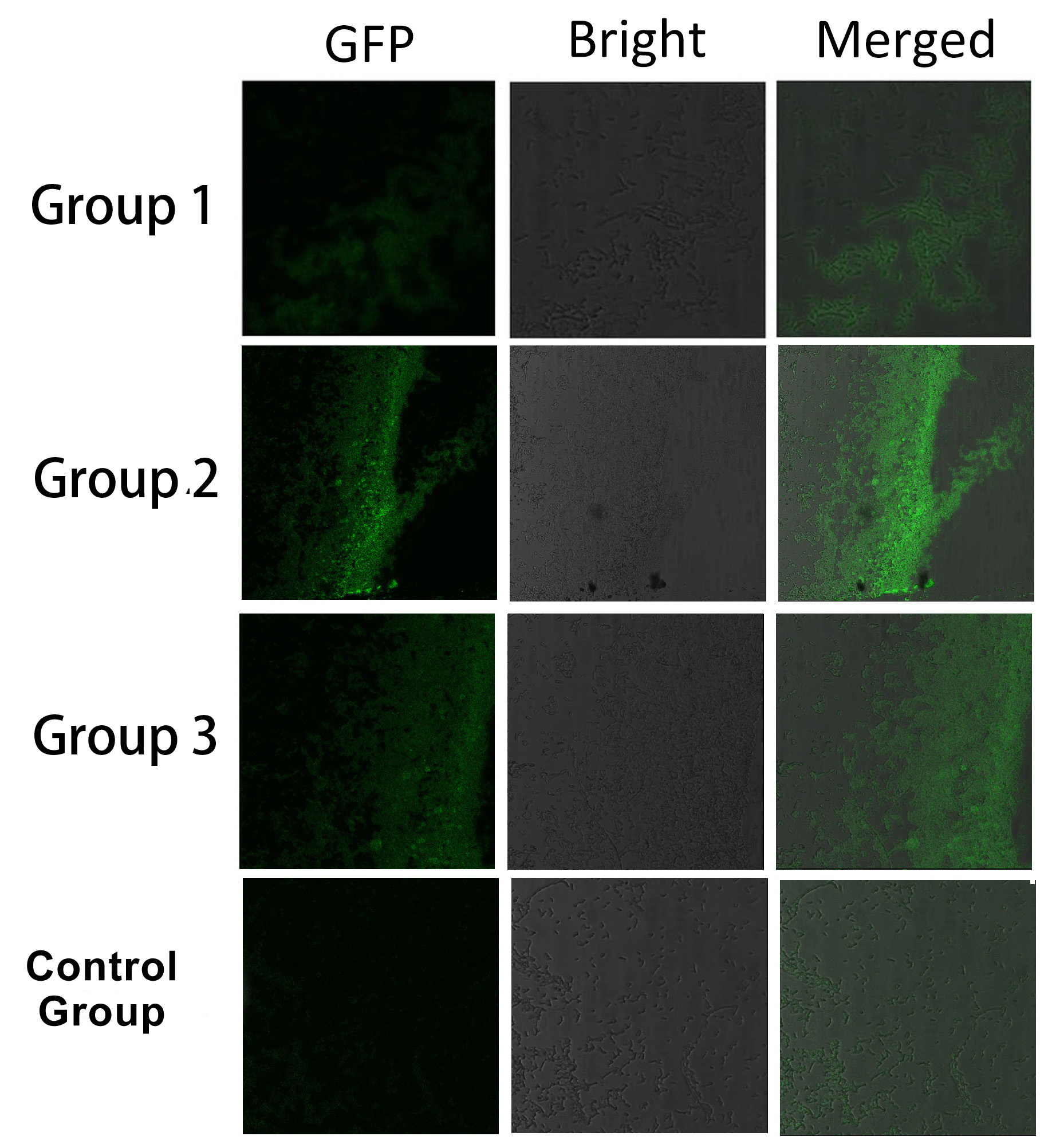 Fig.5 :Bacteria in Group 1, 2 and 3 all show significantly stronger fluorescence intensity than Control Group. So we can conclude that through recruiting interacting domain and ligand, we can assemble proteins into a complex Thus, it is proved that membrane proteins with interacting proteins could interact and dimerize with each other. Thus, by recruiting E.coli native membrane protein Lgt, interacting proteins (SH3, PDZ, GBD) and downstream enzymes, we can easily build a Membrane Accelerator.
Reference
1.Dueber, J. E., G. C. Wu, et al. (2009). "Synthetic protein scaffolds provide modular control over metabolic flux." Nature biotechnology 27(8): 753-759.
2.Valencia-Burton, M., R. M. McCullough, et al. (2007). "RNA visualization in live bacterial cells using fluorescent protein complementation." Nature Methods 4(5): 421-427.
|