Team:Chalmers-Gothenburg/Results
From 2012.igem.org
Contents |
Results Summary
The gene CWP2, encoding a mannoprotein in the cell wall was successfully deleted in the yeast strain IMFD-73. The deletion was confirmed by verification PCR. In addition, a lyticase assay showed that [IMFD-73 Δcwp2::kanMX] was degraded faster than IMFD-73 and this leads us to the conclusion that the cell wall is somewhat weakened in the strains with the deleted cell wall mannoprotein CWP2.
Two different plasmids both containing the receptor gene LHCGR but one with the human signal peptide and the other one with a yeast signal peptide was created. Both plasmids were successfully cloned into IMFD-73 and [IMFD-73 Δcwp2::kanMX] but in neither strains could the receptor be proved as functional in detection of hCG.
The Indigo group managed to obtain different colour changes in different growth media by introducing a plasmid containing the genes tnaA and fmo. For instance, blue bubbles could be seen in one medium and other media turned into different shades of brown. However, we did not manage to determine which compound or reaction that was responsible for the colourful bubbles or colour changes.
[IMFD-73 Δcwp2::kanMX] was successfully transformed with both receptor genes and genes required for bio-indigo production. However, the system was never functional as a pregnancy test kit since it did not give any significant response in the presence of hCG.
More detailed analysis of the results can be found in the sections below.
Survival of yeast in urine
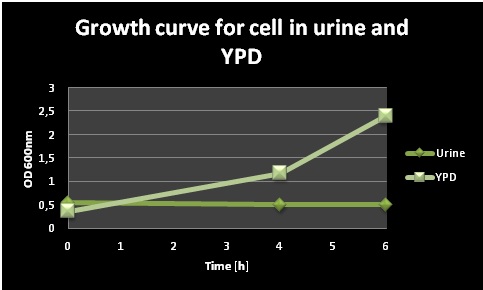
An initial test was performed in order to test the survival of yeast cells in urine, which is an important property of our biosensor if it should function as a pregnancy test. Pre-cultures of a normal lab yeast strain were prepared in 5ml YPD and let grown O/N at 30 °C. The day after, the cells were centrifuged, resuspended in 40 ml YPD and 40 ml filter sterilized urine respectively and grown in shake flasks at 37°C. OD600 measurements were taken at different points of time during the day, and the values can be seen in Figure 1. The cells in urine did not grow; the OD remained the same during the whole day, while the cells in YPD grew very well.
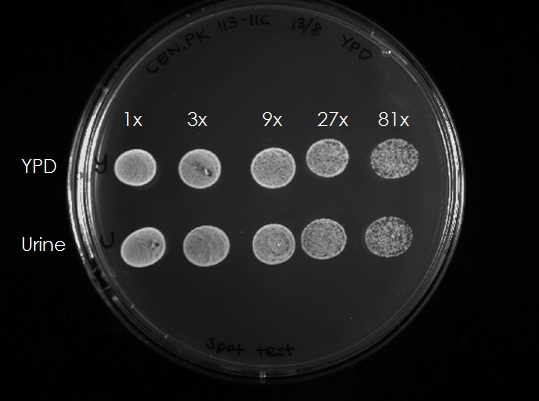
In order to test if the cells still are alive in the urine medium, 300 µl of each culture (after 4h) were taken and OD was adjusted to 0.5. The samples were diluted 3x, 9x, 27x and 81x and 10 µl aliquots of each dilution were spotted on an YPD plate (Figure 2). The cells from both the YPD and the urine media could grow, this means that the cells survived 4h in urine and grew normal after being spotted on a YPD plate. Summarizing, Yeast cells are unable to bud/divide in the urine medium but survive under these conditions.
Expression of human LH/CG receptor
Introduction of indigo synthesizing genes
Deletion of CWP2 gene
One task of our iGEM project was the deletion of the CWP2 gene, which is encoding a cell wall mannoprotein. By removing it, we aimed for higher cell wall permeability and thus higher chances of our ligand to pass the cell wall and to bind to the membrane-bound receptor. The gene deletion was performed according to the bipartite method. The results from the first PCR reactions, where we amplified the overlapping fragments, can be seen in Figure X.
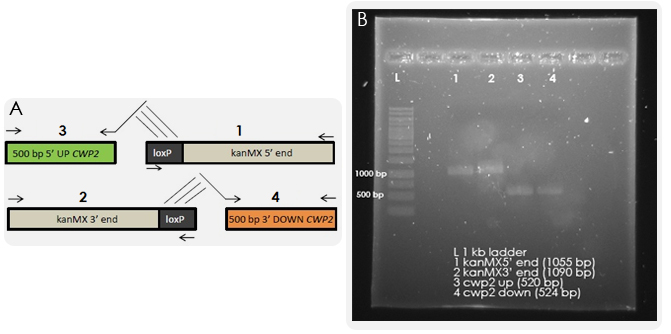
The next step was to fuse together the overlapping fragments. The results of the Fusion PCR are shown in Figure Y. When yeast is transformed with these fragments, homologous recombination should occur which will lead to the exchange of the CWP2 gene with the kanMX cassette.
 "
"