Team:WashU/mobile/Week4
From 2012.igem.org
(Created page with "{{Washuback}} __NOTOC__ ==Website== Once again, the website has undergone a series of changes, such as the new sitemap and the improved background page. A lot of the web pages we...")
Newer edit →
Revision as of 21:42, 25 July 2012
Website
Once again, the website has undergone a series of changes, such as the new sitemap and the improved background page. A lot of the web pages were polished further, and two new Prezis were created to describe our projects. The team is also working on a logo at this point in time.
YLC
The team has finished up the PowerPoint to be presented on the day of the YLC Outreach. In addition, the team made plates as demos for the children using CFP, YFP, GFP and RFP.
Unfortunately, most of the plates had little to no growth. The vectors used were Kan-resistant while the plates were Amp plates. We discovered that one of the RFP vials was incorrect and not properly bricked. It was only a vector for Kan/Amp resistance and did not contain a functioning RFP. We re-plated that day on Kan plates and obtained successful growth of the GFP and RFP (from a different vial). We concluded from this that the only successful construct was GFP and that RFP was a self-ligated J23100 promoter vector.
The following day at our meeting with our advisors, we went over the procedures. Andrew discovered that ligase used was 3 years expired so we threw it out and started use of our new ligase from NEB. We were advised to try gel purification after digestion to try to prevent self-ligation. We also feared that the miniprep might have been done poorly for GFP, YFP, and mCherry since they were done by new guys to the task and poorly harvested. Thus, these were grown in liquid culture and re-miniprepped.
The promoter was also re-examined, and it was determined that we should switch to [http://partsregistry.org/Part:BBa_J23119 J23119], the strongest promoter of the family. We had previously used J23100, a constitutive promoter of the same family, but much weaker than J23119. This DNA was rehydrated on the plate and prepared to clone over Friday night.
To test whether we had the proper constructs for our different fluorescent proteins, the team ran a gel of the digests, shown below.
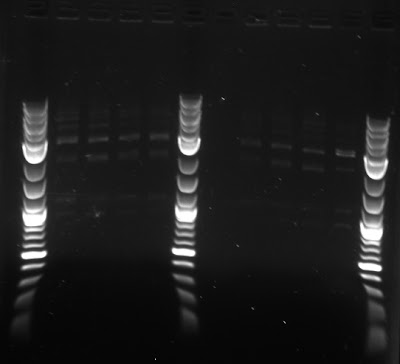
Synechocystis
We plated a Kanamycin resistant synechocystis on plates made with noble agar and glucose, noble agar and no glucose, bacteriological agar and glucose, and bacteriological agar and no glucose.
|
| |
| Result of agar test after four days. Notation: N=noble agar, B=bacteriological agar, -G=without glucose (autotrophic) and +G=with glucose(mixotrophic) |
E. coli
The transformations from last week seemed to have worked. The CFP is extremely light and looks almost like GFP. This could be the result of our UV light, which is quite weak.
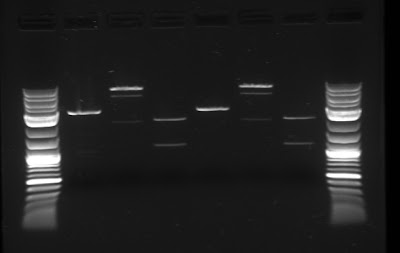
The team has continued to attempt to biobrick together the necessary genes that create the carotenoid pathway in E. coli (see last week for more information). To do this, the team followed the biobrick assembly protocol once more, synthesizing the upstream, downstream and plasmid parts, and then connected them. In order to increase the efficiency and yield of this assembly, gel purification was used to isolate the plasmid for the biobrick assembly. Below is the gel with the upstream, downstream and plasmid parts.
Modeling
We have been hard at work learning what flux balance analysis is all about. We were able to find a recently published genome scale model for Synechocystis PCC 6803, but were unsure how to modify the excel spreadsheet for use with MATLAB. Because of this uncertainty, we decided that it would be best to try and design our own simplified model for Synechocystis that would encompass the carotenoid biosynthesis and all other relevant pathways. At the same time we worked on writing code in MATLAB to parse the data from the excel file into a workable stoichiometric matrix.

 "
"

