Team:Arizona State/Notebook
From 2012.igem.org
(Difference between revisions)
Rohit Rajan (Talk | contribs) (→Water Sample Testing) |
Rohit Rajan (Talk | contribs) (→Water Sample Testing) |
||
| Line 958: | Line 958: | ||
[[File:ASUiGEM_WaterSampleB10232012.jpg|200px]] | [[File:ASUiGEM_WaterSampleB10232012.jpg|200px]] | ||
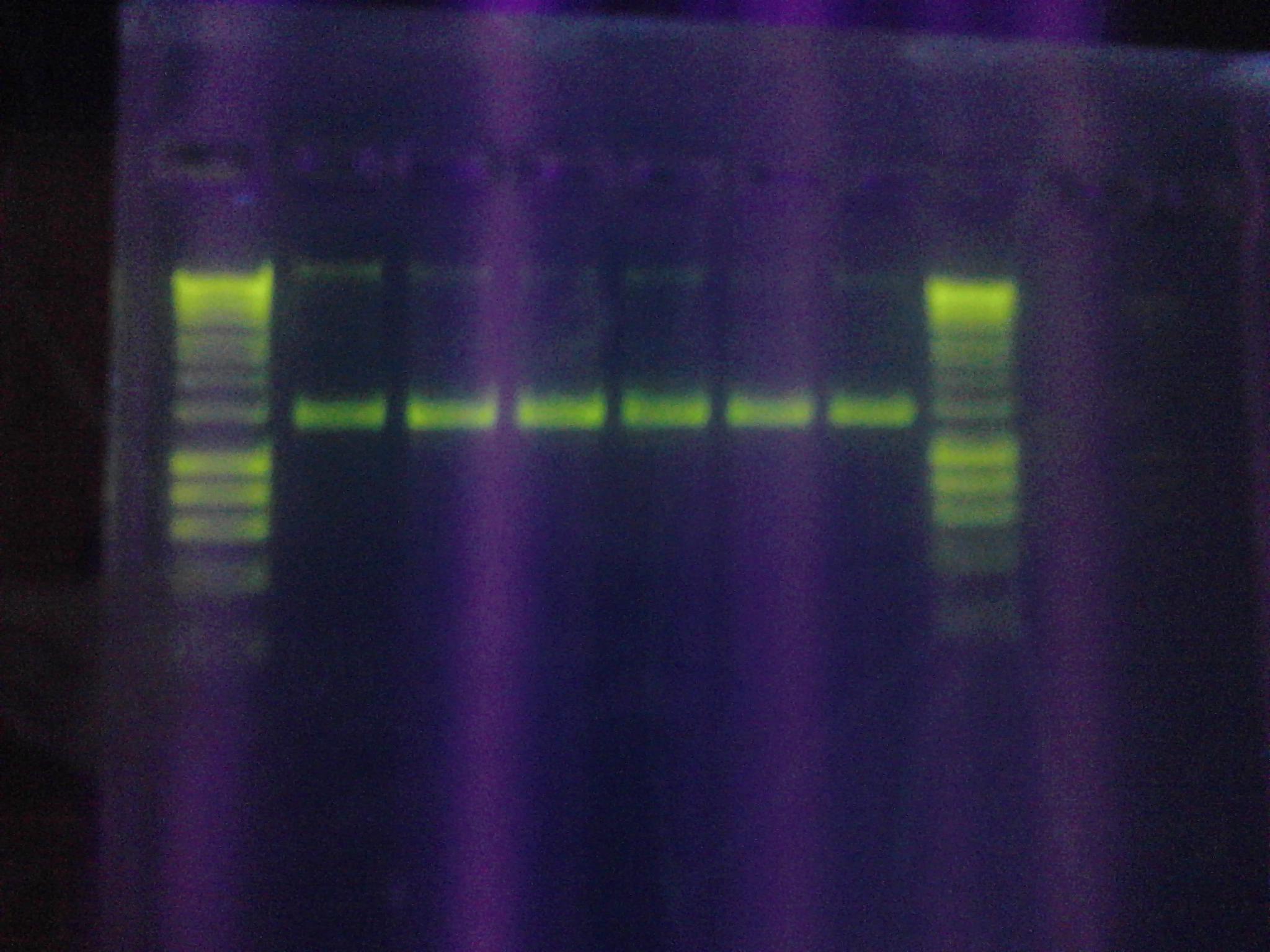
* Figure 3: Control Samples. Left to right, water sample from Artisan Courtyard fountain (ASU campus), water sample from Tempe Town Lake and Nuclease free water. The following picture was taken 10 minutes after X-gal, NEB10 B cell lysate (contains omega fragment of LacZ) and DTT were added to the solution. The following test samples were under buffering conditions. No color change was observed. | * Figure 3: Control Samples. Left to right, water sample from Artisan Courtyard fountain (ASU campus), water sample from Tempe Town Lake and Nuclease free water. The following picture was taken 10 minutes after X-gal, NEB10 B cell lysate (contains omega fragment of LacZ) and DTT were added to the solution. The following test samples were under buffering conditions. No color change was observed. | ||
| - | [[File: | + | [[File:ASUiGEM_WaterSampleC10232012.jpg|200px]] |
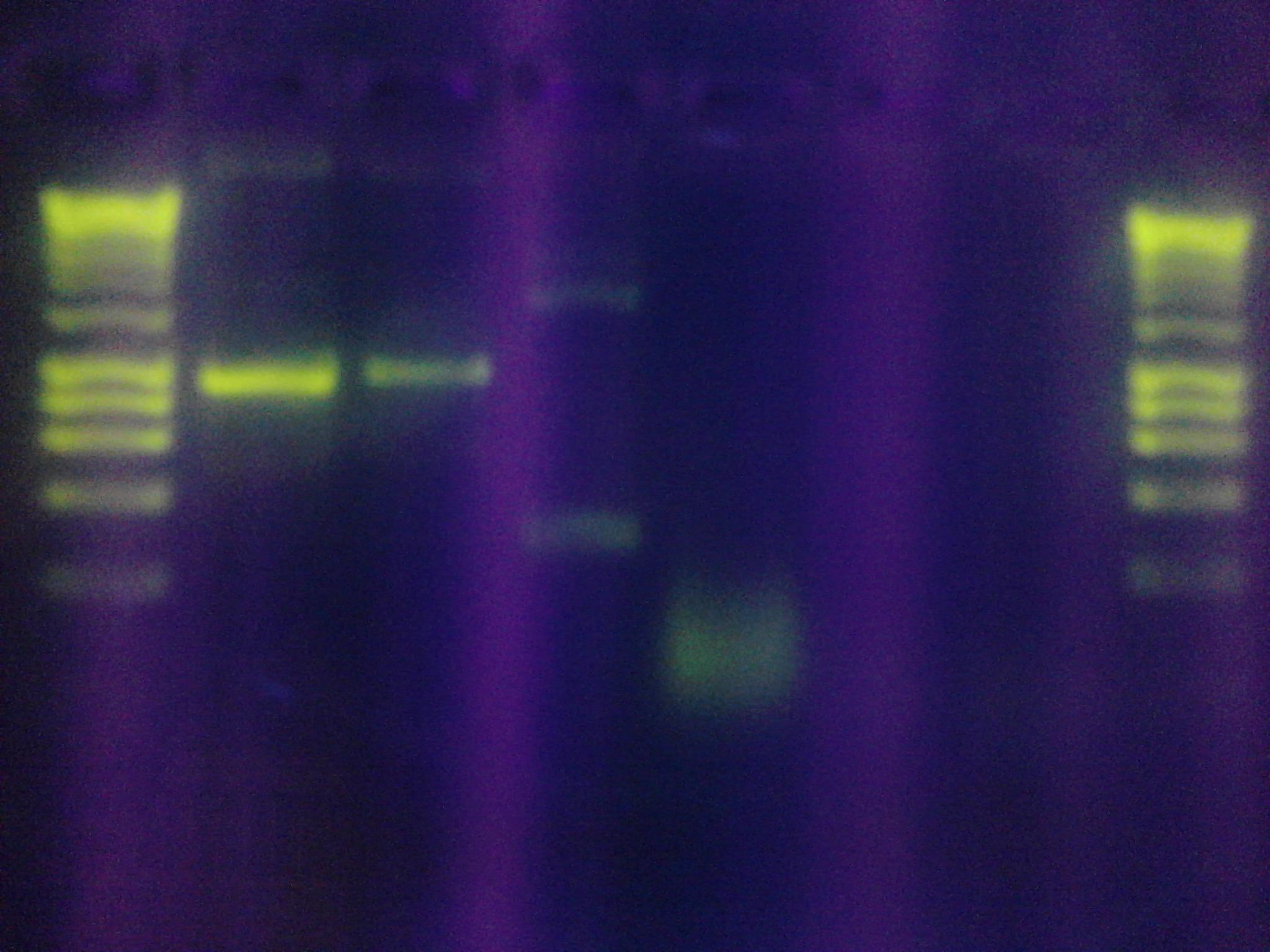
* Figure 4: Left to right, water sample from Artisan Courtyard fountain (ASU campus), Nuclease free water and water sample from Tempe Town Lake. The following picture was taken 10 minutes after X-gal, commercial beta gal enzyme and DTT were added to the solution. The following test samples were under buffering conditions. No color change was observed. | * Figure 4: Left to right, water sample from Artisan Courtyard fountain (ASU campus), Nuclease free water and water sample from Tempe Town Lake. The following picture was taken 10 minutes after X-gal, commercial beta gal enzyme and DTT were added to the solution. The following test samples were under buffering conditions. No color change was observed. | ||
NEED PICTURE | NEED PICTURE | ||
Revision as of 02:23, 27 October 2012
June 07
Strep
- Transformation (LSE)
- Transformed DNA:
- lacZ (well 4:12G, I732019)
- p + lacO (well 1:6G, R0011)
- Cells: neb10beta (donated)
- Protocol from: http://www.neb.com/nebecomm/products/productc3019.asp
- Controls: puc19, no DNA (8 plates)
- Transformed DNA:
June 08
Strep
- Transformation results
- puc19: growth
- negative control: no growth
- lacZ, lacO: possible small colonies
- liquid culture in amp media (100 ug / ml):
- no growth of lacZ, lacO
- growth of puc19
June 12
General
- DH5a Competent Cell Prep
- Streak plated cells on LB no amp plate, let grow overnight
June 13
Strep
- Transformation (LSE)
- Transformed DNA:
- lacZ (well 4:12G, I732019)
- p + lacO (well 1:6G, R0011)
- Cells: DH5 alpha (donated)
- Protocol from: http://openwetware.org/wiki/Haynes:Assembly101 (30 minute transformation)
- Controls: puc19, no DNA (8 plates)
- Transformed DNA:
General
- DH5a Chemically Competent cell prep
- Grew 2 seed colonies from streak plate in LB no amp
- Grew controls to test for contamination
- Both Seed colonies grew, no contamination present
June 14
General
- Competent cell prep
- Prepared CaCl2 buffer solution and CaCl2 glycerol buffer solution
- Grew seed colony in 400mL LB no amp
June 15
General
- Competent cell prep
- Centrifuged falcon test tubes containing liquid colonies
- Resuspended in CaCl2 buffer solution and incubated for 15 mins
- Centrifuged and resuspended in CaCl2 glycerol buffer solution
- Chilled overnight
June 16
General
- Competent cell prep
- Aliquotted 200uL into test tubes
- Stored in -80C
June 17
General
- Streak plated prepared competent cells on LB no amp plate
- Colonies observed
- No picture
June 18
Strep
- Digest: Lac Z (X and P) and p + lacO (S and P)
- Protocol: http://openwetware.org/wiki/Haynes:Assembly101
- Gel extraction - Sigma Aldrich Gel Kit
June 19
Strep
- Ligation
- P+lacO+Lac Z (samples still need to be tested)
- Control (No DNA)
General
- Transformation (LSE)
- Transformed DNA:
- T7 promoter BBa_I712074
- Constitutive promoter BBa_J23102
- Cells: DH5 alpha (donated)
- Protocol from: http://tools.invitrogen.com/content/sfs/manuals/subcloningefficiencydh5alpha_man.pdf (30 minute transformation)
- Controls: puc19, no DNA
- Plated 2 copies of each (100 ul, 250 ul) on LB amp plates.
- Transformed DNA:
- Made 50 LB Amp plates.
June 20
Strep
- PCR
- Lac Z
- Primers: Standard BBa F and R primers (http://partsregistry.org/Primers/Catalog)
- 1000 fold dilution DNA used
- Protocol
- PCR water - 74.4 uL
- 1000 fold dilution DNA - 6 uL
- dNTP (10mM) - 2.4 uL
- F and R primers (10 uM) - 6 uL each
- 5x HF buffer - 24 uL
- Phusion DNA polymerase - 1.2 uL
- Lac Z
- Digest
- P+Lac O
- S and P
- Treated with Antartic Phosphatase (avoids religation)
- P+Lac O
- Plated negative control on LB Amp plate
- Liquid cultures of T7 promoter and constitutive promoter
General
- Transformation (LSE)
- Transformed DNA:
- RBS (well 1:1H BBa_B0030)
- TetR GFP (well 2:8A Part:BBa_I13522)
- Cells: DH5 alpha (donated)
- Protocol from: http://tools.invitrogen.com/content/sfs/manuals/subcloningefficiencydh5alpha_man.pdf (30 minute transformation)
- Controls: puc19, no DNA
- Transformed DNA:
June 21
General
- Made Liquid Cultures of E.coli transformed with RBS B0030
- Made Liquid Cultures of E.coli transformed with TetR GFP
- miniprepped and nanodropped T7 promoter BBa_I712074 and Constitutive promoter BBa_J23102 liquid cultures
- liquid cultures:
- RBS1
- RBS2 (duplicate_
- GFP1
- puc19
- negative controls
- 5 ml LB amp
- overnight cultures
- replated GFP1 & 2 (duplicates)
- Nanodropped plasmid DNA samples
- Constitutive promoter 1: 2.554 ng/uL
- Constitutive promoter 2: 2.345 ng/uL
- T7 promoter 1: 3.369 ng/uL
- T7 promoter 2: 3.049 ng/uL
June 22
Strep
- PCR clean up (Sigma Aldrich Kit) Lac Z samples
- Digest Lac Z with X and P
- Nano Drop the PCR clean samples:
- Lac O: 2.554 ug/uL
- Lac Z: 3.369 ug/uL
- Gel
- PCR
- Lac Z (25ul rxn)
- Primers: Standard BBa F and R primers (http://partsregistry.org/Primers/Catalog)
- Varied Concentration DNA samples
- Syzygy Mean Green MM
- Lac Z samples - 10 fold D, 100 fold D, 1000 fold D and Regular DNA.
- Lac Z (25ul rxn)
- PCR cleaned samples
General
- Miniprepped liquid cultures: RBS (well 1:1H BBa_B0030) and TetR GFP (well 2:8A Part:BBa_I13522)
- Picked colonies:
- 1 colony from double terminator (dt1) plate
- 1 colony from t7 polymerase (pol1) plate
- 1 colony from puc19 plate (positive control)
- 1 colony from dh5a plate (negative control)
- started liquid cultures of each colony (5 mL LB amp each)
June 26
General
- Transformation:
- Transformed DNA:
- double terminator (B0017, 2:6K)
- T7 RNA polymerase (I715038, 2:15C)
- puc19, negative control
- Protocol from: http://tools.invitrogen.com/content/sfs/manuals/subcloningefficiencydh5alpha_man.pdf (30 minute transformation)
- Cells: dh5a
- Transformed DNA:
June 27
General
- 6-26 transformation results:
- Controls correct
- 2x terminator: ~19 colonies
- RNA pol: 1 colony
- Liquid cultures including controls
June 28
General
- Miniprepped double terminator (B0017, 2:6K) and T7 RNA polymerase (I715038, 2:15C) liquid cultures
July 2
General
- Cleaned up liquid waste
- Made SOB media
Magainin
- Finalized oligos for magainin construct
July 3
General
- Third attempt of putting the bricks together. And samples were plated on X-gal plates to show that the construct worked.
- Autoclaved SOB media
- Added glucose to make SOC media
- Nanodropped double terminator (B0017, 2:6K) [DT1: 24.5, DT2: 29.6] and T7 RNA polymerase (I715038, 2:15C) [P1: 64.6, P2: 55.3] liquid cultures
July 12
Magainin
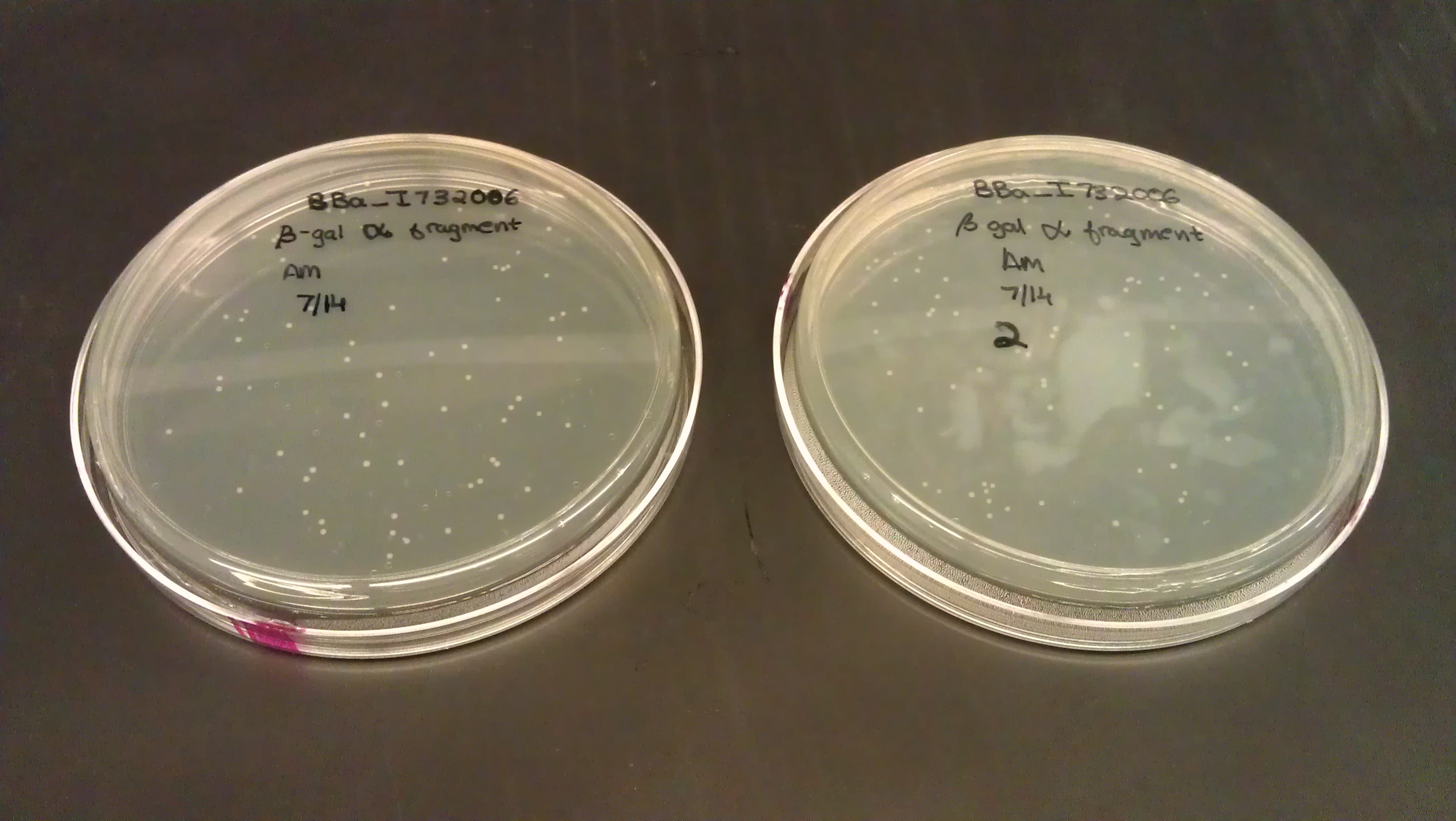
- Searched for split beta gal fragment. Alpha fragment (BBa_I732006) was found on the distribution plates and transformed (NEB 10B).
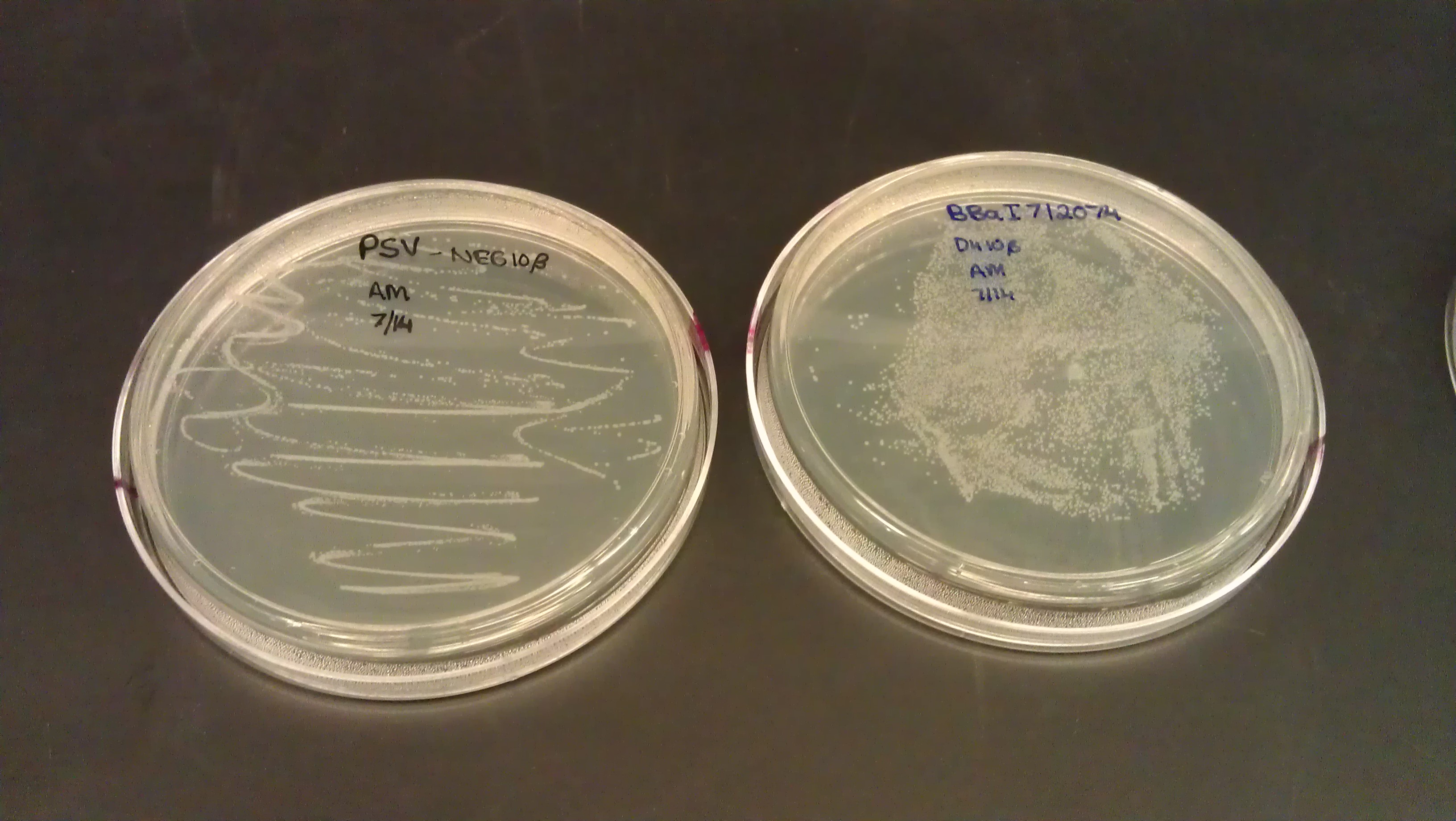
July 14
Magainin
- Designed primers to remove the stop codon on the alpha fragment. Also attained two flexible linkers from Dr. Haynes and transformed them.
July 15
Magainin
- Ran a miniprep of Bgal alpha 1&2, T7 and PSV
- Procedure from GenElute HP PLasmid Miniprep Kit (Sigma-Aldrich)
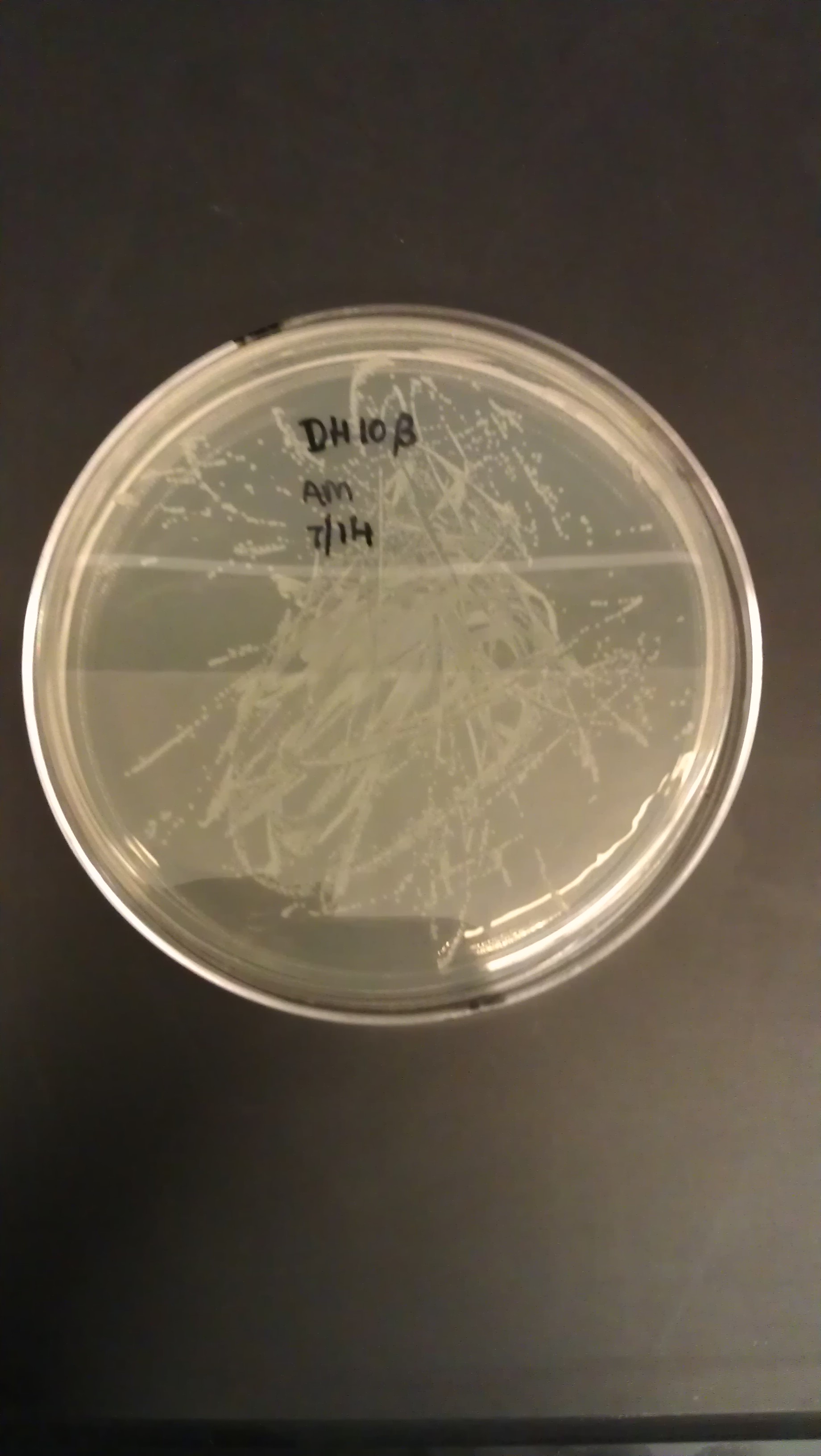
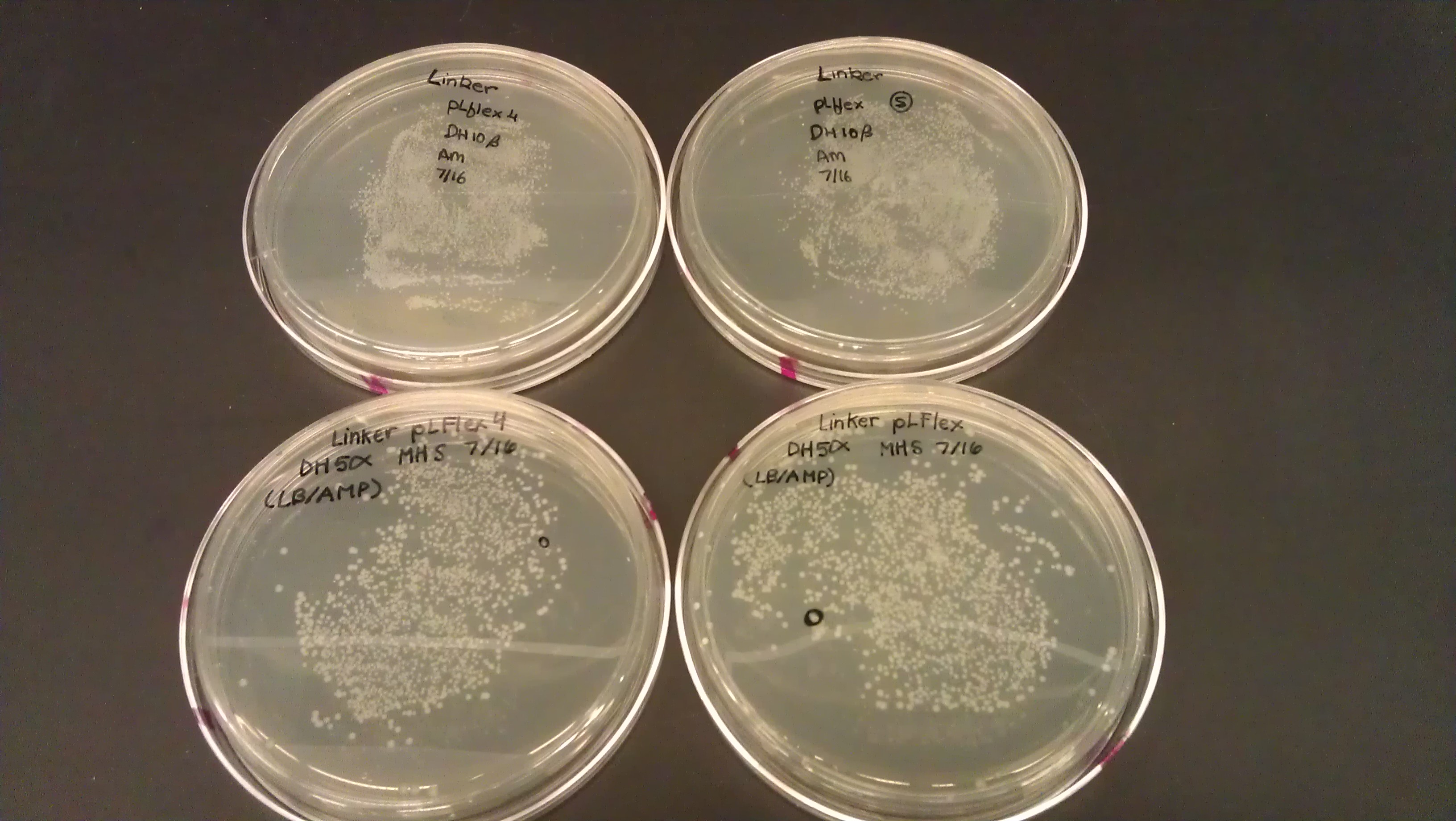
July 16
Strep
- Transformation of PLflex (part: BBa_J176040) and PLflex4 (part: BBa_J176130) linkers.
- Incubate at 37C for 8 hours for DH 5alpha turbo cells
- Inoculated 5 mL LB/AMP media in 15 mL culture tubes and added the colony. Incubated in 37C overnight.
July 17
Strep
- Miniprep of pLFlex DH5alpha and pLFlex 4 DH5alpha
- Procedure comes from Zymo Research's Zyppy Kit.
- Ran the spectrophotometer to determine the concentration for the miniprep DNA for the linker pLFlex and linker pLFlex 4
- Concentrations were:
- pLFlex: 73.792 ng/uL
- pLFlex 4: 38.587 ng/uL
- Absorbance ratios were:
- pLFlex: 1.864
- pLFlex 4: 1.851
- Ran the miniprep for pLFlex and pLFlex 4 again.
- Same procedure
- Ran the spectrophotometer again
- Concentrations were:
- pLFlex: 48.095 ng/uL
- pLFlex 4: 18.214 ng/uL
- Absorbance ratios were:
- pLFlex: 1.79
- pLFlex 4: 1.85
- Transformation was successful but 8-9 hour growth period was not sufficient for DH5 alpha.
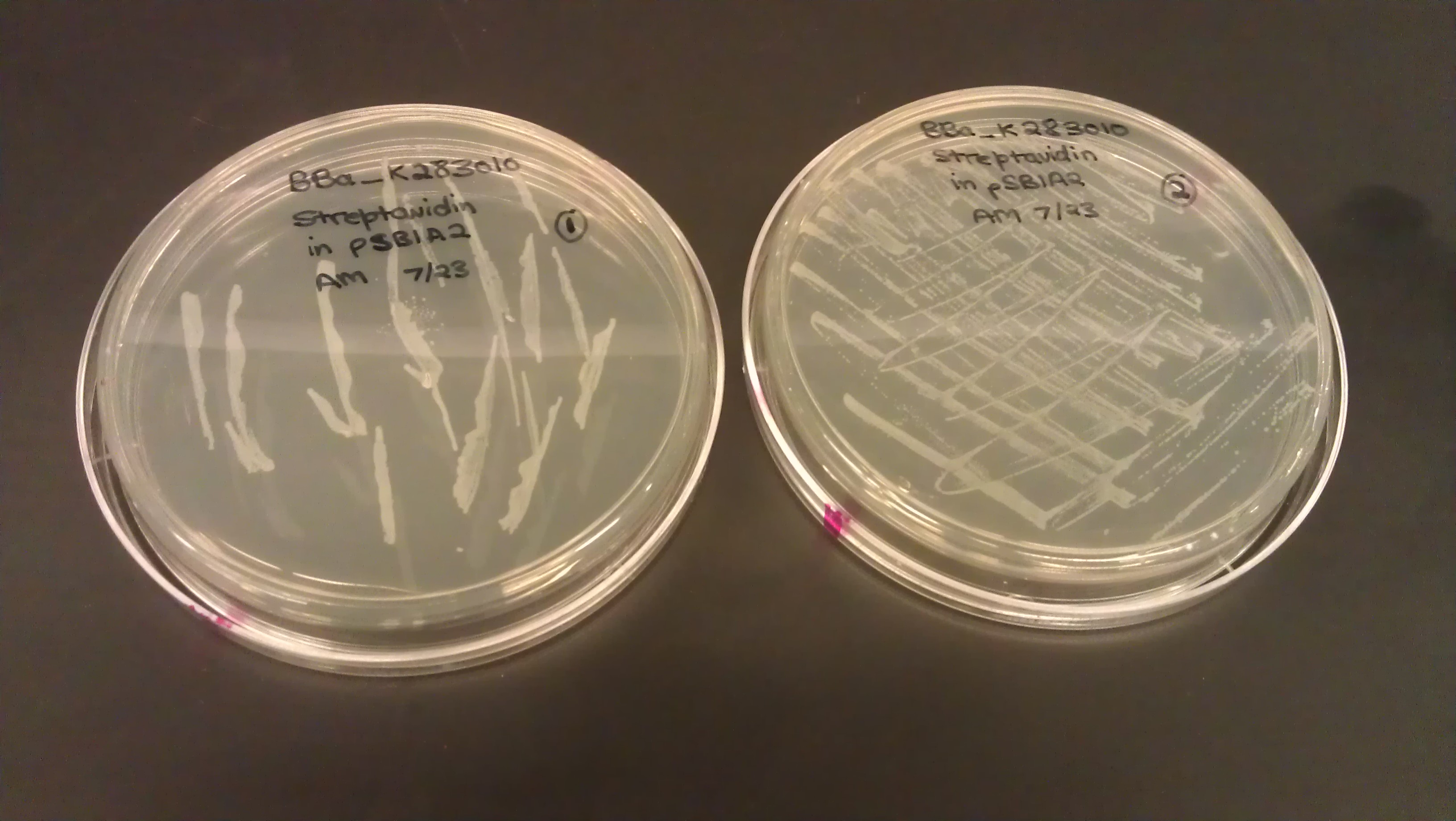
July 23
Strep
- Plated BBa_K283010 (Streptavidin) on LB amp plate from the agar stab provided from the iGEM head-quarters.
- Incubated the dish overnight at 37 C.
- Plate 1: pipet tip, colonies are in wells, neither distinct nor segregated
- Plate 2: inoculating loop, numerous non-distinct colonies on the plate
July 24
Topo
- Plasmids arrived courtesy of University of Pennsylvania School of Medicine
- pET29a vectors containing coding sequence for Topoisomerase mutants CSCS and CSCS D168A described here
July 25
Strep
- Ran a miniprep of streptavidin BBa_K283010
- Protocol from Zymo Research's Zyppy Plasmid Miniprep Kit
Topo
- Tranformed CSCS topo 0 plasmid and CSCS D168A topo into DH5a Turbo cells (with neg control and Puc19 neg control)
- Plated on Kanamycin plates
July 26
Topo
- Picked colonies colonies from Topo 0 and Topo D168A and grew liquid cultures in Kan media
July 27
Topo
- Miniprepped liquid colonies and nanodropped.
- Plasmid concentrations
- Topo O: 63.7 ng/uL
- Topo D168A1: 125.4 ng/uL
- Topo D168A2: 116.6 ng/uL
July 30
General
- Prepared Kan Media and Kan Plates
July 31
Topo
- PCR amplified polylinker sequence of Topo plasmid with Promega GoTaq protocol
- Used pET29a upstream forward primer and T7 terminator reverse primer
August 3
Topo
- Submitted pET29a Topoisomerase plasmid to Biodesign for sequencing
- Resuspended GFPT1 and GFPT2 oligos with molecular grade (nuclease-free) H2O.
- Final Concentration 100uM
- (gfpt1 top1, gfpt2 top1, gfpt1 top2, gftp2 top2, gfpt1 bot1, gfpt2 bot1, gfpt1 bot2, gfpt2 bot2)
- (3uL of each oligo + 2uL 10x annealing buffer, 6uL molecular grade H2O. 20uL Reactions)
- Final Concentration 100uM
- Heated for 5 minutes at 100C. Let cool to room temperature on the heating block, stored at -20C.
- Digested BBa_I13522 with XbaI and PstI.
- Attempted ligating annealed oligos into a digested plasmid from Ryan (realized it was cut with E and P).
August 6
Topo
- Annealed oligos for GFPT1 and GFPT2 (target probes)
- Ligated oligos with digested GFP plasmid (BBa_I13522)
- Transformed into competent DH5alpha
- Added SOC and incubated at 37C for 15 minutes.
- Plated on amp treated plates.
August 7
Topo
- Only one colony on each plate (both were white)
- Picked colonies and started 5mL LB amp cultures of each, stored at 37C
- Stored plates in 37C
August 8
Topo
- Picked the colonies again and started new liquid cultures (5mL LB amp).
- Discarded cultures for 8/7/12
August 9
Topo
- Miniprepped 3mL of each 8/8/12 culture and nanodropped:
- gfpt1 - 155 ng/uL
- gfpt2 - 114 ng/uL
- Digested gfpt1 and gfpt2 with X and P
- Ran on a 1% agarose gel with the digested GFP plasmid and hyperladder V
- Didn't work
- Made glyercol stocks with aliquot of the remaining liquid cultures
August 10
Magainin
- PCR of Alpha-4, 1-omega, omega, and alpha fragments using corrected primers
August 13
Magainin
- Ran gel of split beta gal fragments. Confirmed 3 out of the 4 fragments except for the alpha-4 fragment.
Topo
- Prepared sequencing samples
- Sample w/ Primer:
- GFPT1 w/ VF2 GFPT1 w/ VR GFPT2 w/ VF2 GFPT2 w/ VR
- 200ng of DNA + 16 pmol of primer
- Annealed oligos again GFPT1/2
- Repeated ligation of oligos with digested GFP plasmid (BBa_I13522)
- Followed Haynes assembly protocol instead of standard DH5alpha protocol. (http://openwetware.org/wiki/Haynes:Assembly101)
- Transformed ligations into competent DH5alpha
- Plated on amp treated plates
August 14
Topo
- Took pictures of plates
- Green-white screened plates
- Picked 4 white colonies from each of gfpt1/2 plates
- Made 5mL LB amp cultures of each colony
- Delivered GFPT1/2 dna samples to biodesign for sequencing (samples from 8/13/12)
Magainin
- Assembled magainin insert via Overlapping oligo assembly
- Digested pUC 19 plasmid with EcoRI and PstI
- Transformed Magainin insert into digested pUC 19 plasmid. Failed. Probably too much X-gal on plate.
Ran gel for the beta gal alpha-4 fragment. Failed. Fragment not in the correct size-band.
August 15
Topo
- [http://129.219.2.10/home2/dnalims/fragment/32/21/pfasta/topo0_T7Term.seq Topo 0], [http://129.219.2.10/home2/dnalims/fragment/32/21/pfasta/D168Atopo1_T7Term.seq D168A Topo1], and [http://129.219.2.10/home2/dnalims/fragment/32/21/pfasta/D168Atopo2_T7Term.seq D168A Topo2] sequence results
- Miniprepped 3mL of each liquid culture of GFPT1/2
- Prepared glycerol stocks using 100uL of each liquid culture
- Nanodropped samples:
- GFPT1-1 - 172.6 ng/uL
- GFPT1-2 - 203.7 ng/uL
- GFPT1-3 - 197.4 ng/uL
- GFPT1-4 - 178.9 ng/uL
- GFPT2-1 - 107.3 ng/uL
- GFPT2-2 - 131.2 ng/uL
- GFPT2-3 - 145.5 ng/uL
- GFPT2-4 - 172.0 ng/uL
August 16
Magainin
- Replated magainin insert + plasmid into the grid. Failed. All blue colonies meaning that no insert.
Tried gel for all gel-isolated fragments. Failed. Did not get a band in the 2000 bp region. Only got things below 200 bp.
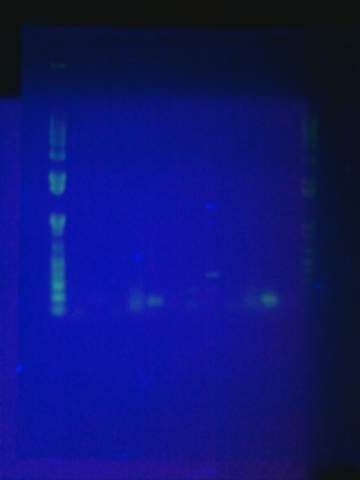
August 17
Topo
- GFPT1 sequence confirmed
- Prepared aliquots of GFPT2 minipreps from 8/15/12 for sequencing
- Delivered GFPT2 samples to biodesign for sequencing
August 18
Strep
- PCR’ed Streptavidin (biobrick) and omega fragment of beta (PSV plasmid) and confirmed size on a gel.
Streptavidin was digested with E and S. Also, both flexible linkers attained from Dr. Haynes were digested with E and X and dephosphorlated.
August 19
Strep
- Restricted PLFlex and PLFlex 4 with ecori and xbai.
- Restricted Bgala with Xbai and psti
- Used Hayne's Lab Protocol
- GFPT2 sequences confirmed
August 22
Strep
- Transformed the two constructs.
- The two constructs were confirmed on gel and were ligated together to attain Streptavidin + pLFlex and Streptavidin+pLFlex4.
August 27
Topo
- Revived GFPT1 (from 8/9/12) and GFPT2 (2-2 from 8/15/12) cultures from glycerol scrapes
- Made 4mL cultures in LB Amp
August 29
Topo
- Discarded GFPT1/2 cultures from 8/27/12
- Revived GFPT1 (from 8/9/12) and GFPT2 (2-2 from 8/15/12) cultures from glycerol scrapes
- Made 4mL cultures in LB Amp
- Digested GFPT1/2 with X and S
- Ran a 1% agarose gel with GFPT1/2 digestions
- Cut out inserts and GFPT2 backbone and stored in 4C for gel extraction and tandum repeat assembly experiments
August 30
Topo
- Prepared extra glyercol stocks of GFPT1/2 cultures from 8/29/12
- Miniprepped 3mL of each culture, stored at -20C
September 2
Strep
- Digested strep+pLFlex4(L4) with S and P and pLflex(L) with E and X and inserted alpha and omega fragments of beta gal.
- Transformed these constructs
September 10
Strep
- Assembled:
- Strep+L4+α which was a success
- Strep+L4+ω which was a success
- Strep+L which was a success
- All digested samples were confirmed on a 1% agarose gel
September 18
Strep
- Assembled:
- Prsf Duest+Strep+L4+α which failed
- Prsf Duet+Strep+L4+ω which failed
- Strep+L+α which was a success
- Strep+L4+ω which was a success
- All digested samples were confirmed on a 1% agarose gel
- No picture of the gel.
September 19
Strep
- Assembled
- Prsf Duest+Strep+L4+α which failed
- Prsf Duest+Strep+L4+ω which failed
- Prsf Duest+Strep+L+α which failed
- Prsf Duest+Strep+L+ω which failed
- All digested samples were confirmed on a 1% agarose gel
Topo
- Set up VF2/VR endpoint PCR for double transform minipreps
- 1-1, 1-1I, 1-2, 1-2I, 1-3, 1-3I, 2-1, 2-1I, 2-2, 2-2I, 2-3, 2-3I, GFPT1 (positive controls), GFPT2 (positive controls)
- Annealing temp set to 55C for 25 cycles
- Resuspended GFPT1 probe and GFPT2 probe oligos in molecular grade H2O (Final concentration: 100uM), stored at -20C
September 21
Strep
- Miniprepped all the streps samples and digested them.
September 22
Strep
- Assembles with new Prsf samples
- Prsf Duest+Strep+L4+α which failed
- Prsf Duest+Strep+L4+ω which failed
- Prsf Duest+Strep+L+α which failed
- Prsf Duest+Strep+L+ω which failed
- Failed because outgrowth step want provided.
- All digested samples were confirmed on a 1% agarose gel
September 25
Topo
- Resuspended pSB1A2 FWD and pSB1A2 REV (amp resistance primers) oligos in molecular grade H2O (Final concentration: 100uM), stored at -20C
- Prepared 1.6uM dilutions (500uL)
- Did endpoint PCR using pSB1A2 primer pair on
- Topo, Topo IPTG, Topo D168A, Topo D168A IPTG, 1-1, 1-1I, 2-1, 2-1I, GFPT1, GFPT2
- Did endpoint PCR using VF2/VR primer pair on
- Topo, Topo IPTG, Topo D168A, Topo D168A IPTG
- Annealing temp 55C for 25 cycles, stored products at -20C
- Made 3mL liquid cultures of the shipping vector (pSB1C3 with RFP insert) in DH5alpha in chloramphenicol resistant LB
- Made 10mL liquid cultures of:
- Topo in kanamycin
- topo D168A in kanamycin
- topo + GFPT1 in kanamycin + ampicillin
- topo + GFPT2 in kanamycin + ampicillin
- stored @ 37C
September 26
Strep
- Assembled
- Prsf Duest+Strep+L4+α which was a Success
- Prsf Duest+Strep+L4+ω which was a Success
- Prsf Duest+Strep+L+α which was a Success
- Prsf Duest+Strep+L+ω which was a Success
Topo
- Picked two new colonies for topo + GFPT2 and grew 10mL cultures of amp+kan LB broth for each
- Ran a gel with Hyperladder I, GFPT1 pSB1A2, GFPT2 pSB1A2, Topo pSB1A2, Topo D168A
- Ran a gel with PCR samples from 9/19 and 9/25 Hyperladder I, 1-1, 1-1I, 2-1, 2-1I, 2-1I(VF), 2-1(VF), 1-1I(VF), 1-1(VF), Hyperladder I
- Samples in wells 2-5 used the pSB1A2 primer pair. Samples in wells 6-9 used VF2/VR primer pair.
- Revived GFPT1/2 from glycerol stocks (from 8/9 and 8/15) in 5mL LB amp each.
- Nanodropped miniprepped DNA samples:
- pSB1C3 I - 253.75 ng/uL
- pSB1C3 II - 258.38 ng/uL
- Topo - 24.84 ng/uL
- Topo I - 17.33 ng/uL
- Topo D168A - 7.24 ng/uL
- Topo D168A I - 15.62 ng/uL
- 1-1 - 16.49 ng/uL
- 1-1I - 16.46 ng/uL
- 2-1 - 142.05 ng/uL
- 2-1I - 16.98 ng/uL
- Picked new colonies from Topo, Topo D168A, Topo + G1, Topo + G2 and made 1mL colonies in their respective medias
- Stored at -37C
September 27
Strep
- Testing R0011 promoter. Digested R0011 with S and P generated the following assembly:
- R0011+Strep+L4+α which failed
- R0011+Strep+L4+ω which failed
- R0011+Strep+L+α which failed
- R0011+Strep+L+ω which was a Success
Topo
- Prepared serial dilutions of GFPT2 plasmid 1:10, 1:100, 1:1000, 1:10000
- Prepared primer mixes for pSB and VF/VR primers
- Prepared realtime PCR plate, ran RT-qPCR with annealing temp 57
- Miniprepped GFPT1/2 cultures from 9/26
- Nanodropped Miniprepped DNA:
- GFPT1 - 276.12 ng/uL
- GFPT2 - 219.84 ng/uL
- Used 100uL of each 1mL culture from 9/26 to seed 10 mL cultures in their respective media
- Added 10uL of 1M IPTG to each culture ~4 hours after seeding
- Removed cells from 37C ~4 hours after IPTG inducing
- Pelleted and lysed following the bugbuster protocol (http://openwetware.org/wiki/User:Behzad_Damadzadeh/Notebook/PcTF_Subcloning_in_E-coli/2012/05/22) (used lysonase for Topo and Topo D168A only)
- HIS purified proteins using the Zymo HIS purification kit
- Digested pSB1C3 plasmid with X and P
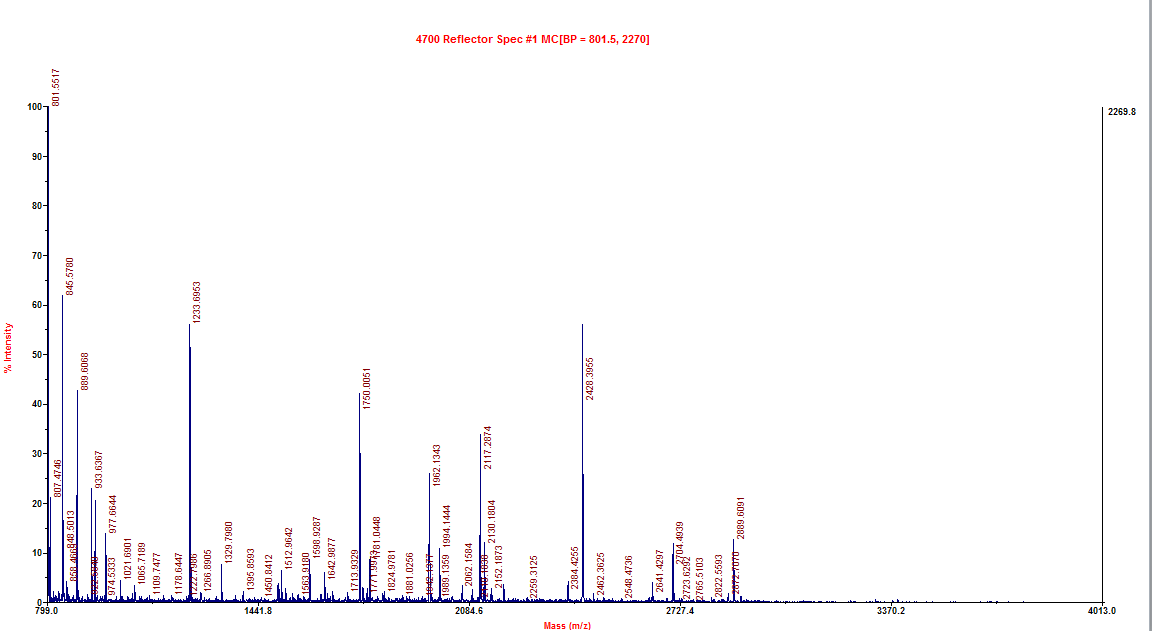
- Sent protein samples (HIS purified samples from 9/19, 20uL) and ssDNA control (GFPT1/2 probe oligos 20uL @ 10uM) for mass spec
September 28
Topo
- Tested another promoter (P2547) for the construct and repeated the assembly from 9/27.
- Resuspended 8 new oligos, final volume 100 uM each
- Annealed pet29 top/bot oligos
- Redid the RT-PCR, 3x primer concentration, added 1:1 plasmid concentration
- Made 1.6uM aliquots of primer stocks 1-7 (including topo add X primer previously ordered)
- Diluted aliquot of Topo D168A plasmid 4:10
- Diluted PSV plasmid 2:20
- Performed Endpoint PCR on:
- Topo D168A using primers 1,2 and primers 1,3
- PSV using primers 4-5 and primers 6-7
- 4 samples with low primer concentration, 4 samples with twice as much primer (labelled 'H')
- Miniprepped Topo1 2, Topo1 3, Topo1 4 (biobricked Topo D168A without T7, multiple colonies from ligation into shipping vector)
- Nanodropped:
- Topo1 2 - 63.63 ng/uL
- Topo1 3 - 75.07 ng/uL
- Topo1 4 - 184.93 ng/uL
- 1mL chloramphenicol cultures of GFPT1/GFPT2 in shipping vector prepared
- Digested Topo1 2, Topo1 3, Topo1 4 with E and P
- Ran digested Topo plasmids on 1% agarose gel with Hyperladder I
- Revived cultures of 2xGFPT1, 2xGFPT2, Topo, and Topo D168A (4 mL cultures each in their respective medias)
- T5 exonuclease treated miniprepped plasmids (1-1, 1-1I)
- A1 1-1I + t5
- B1 1-1 + t5
- A2 1-1I untreated
- B2 1-1 untreated
September 29
Strep
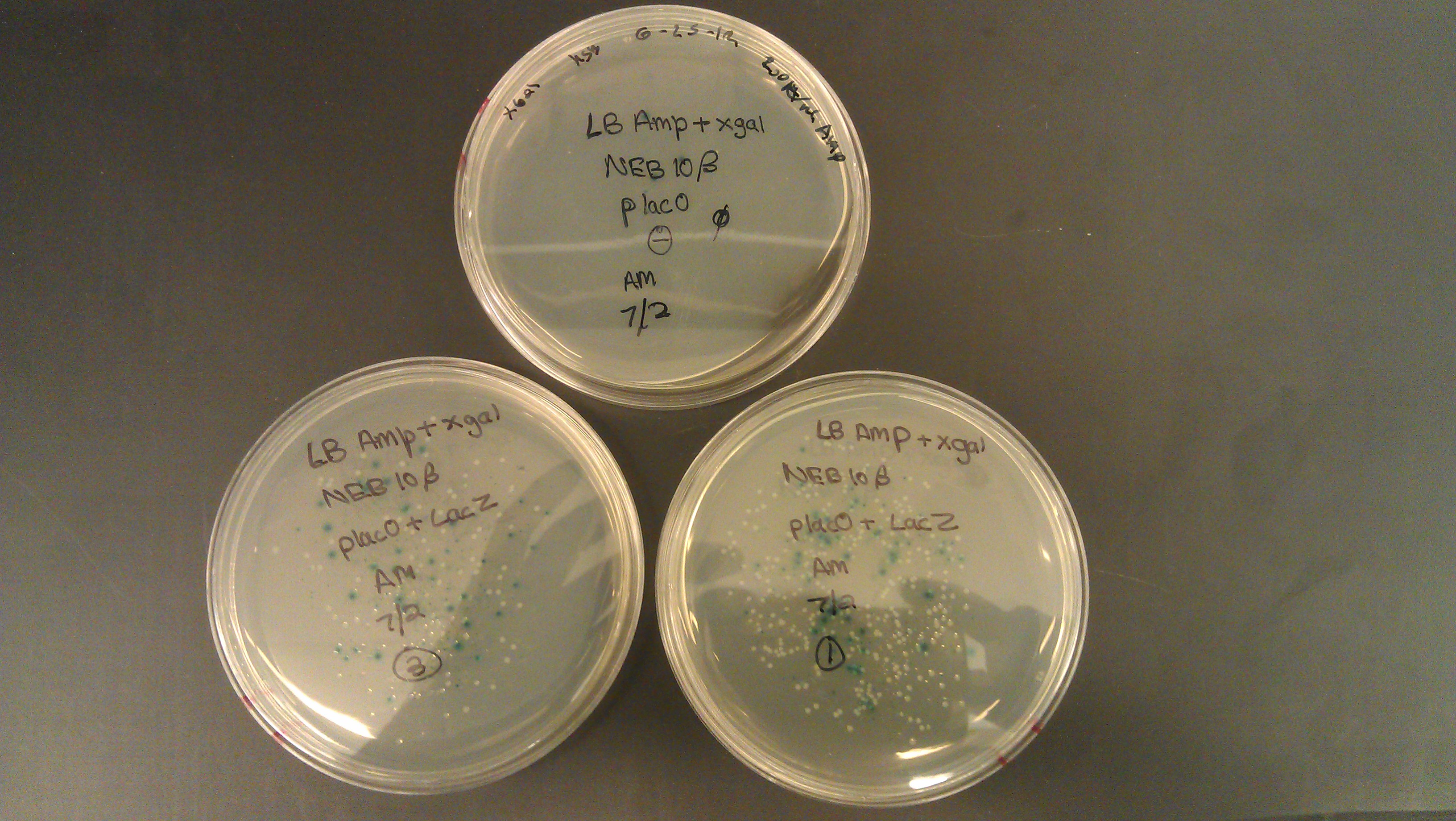
- Assembly was successful as liquid cultures were made. It was realized that due to the lack of beta gal negative strains the omega fragment couldn’t be tested and R0011 wouldn't code for the protein due to the lack of an RBS.
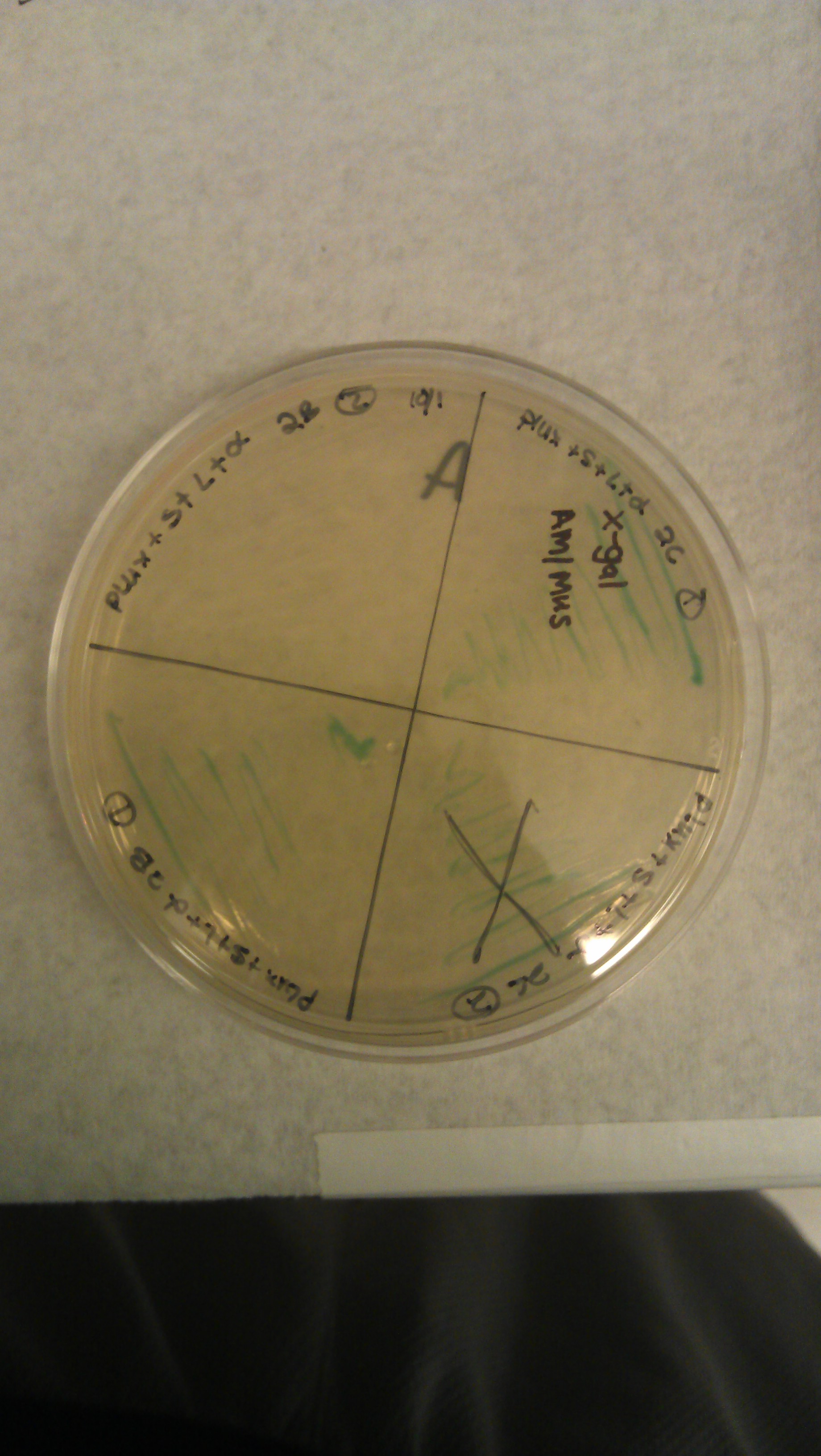
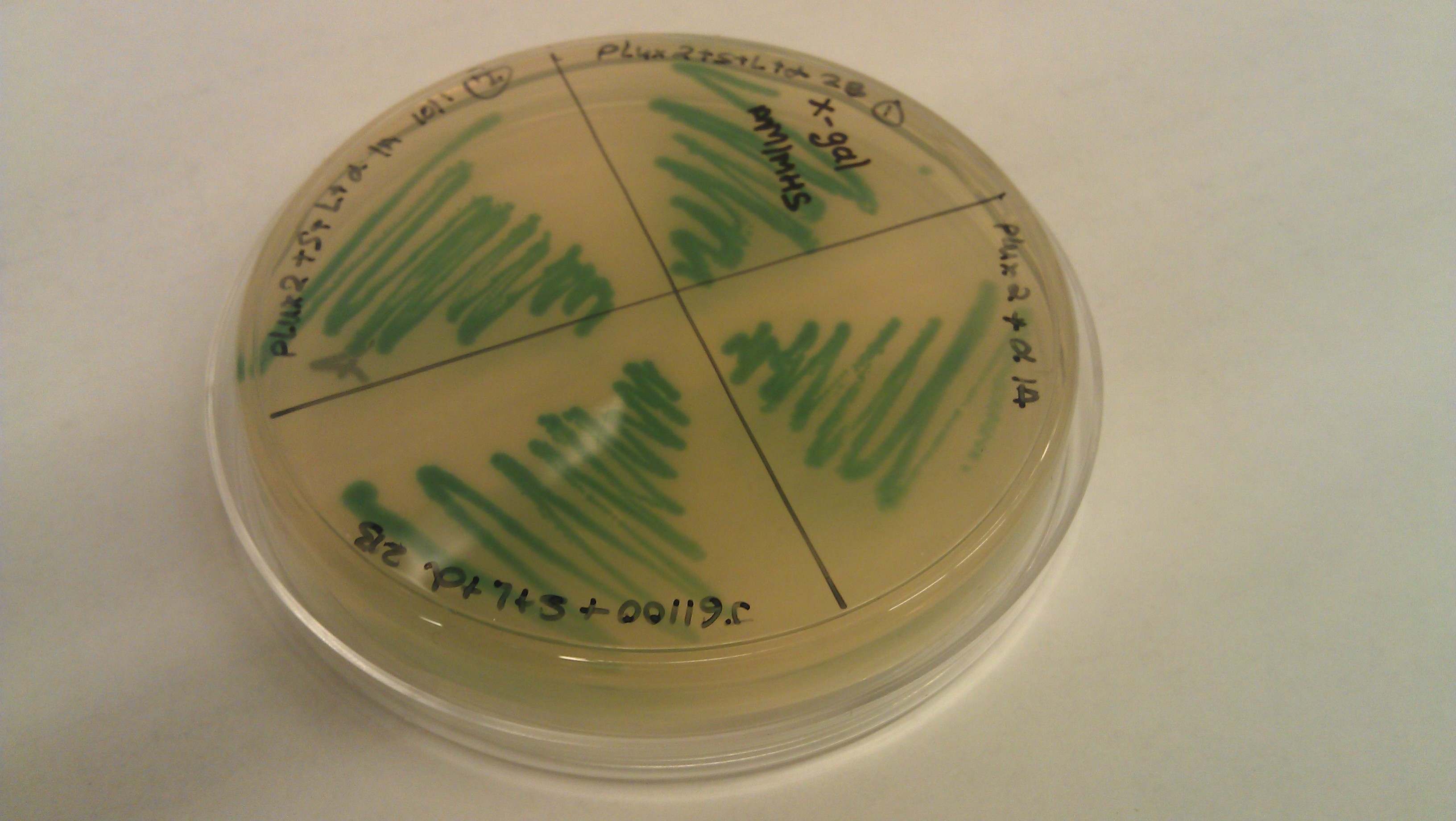
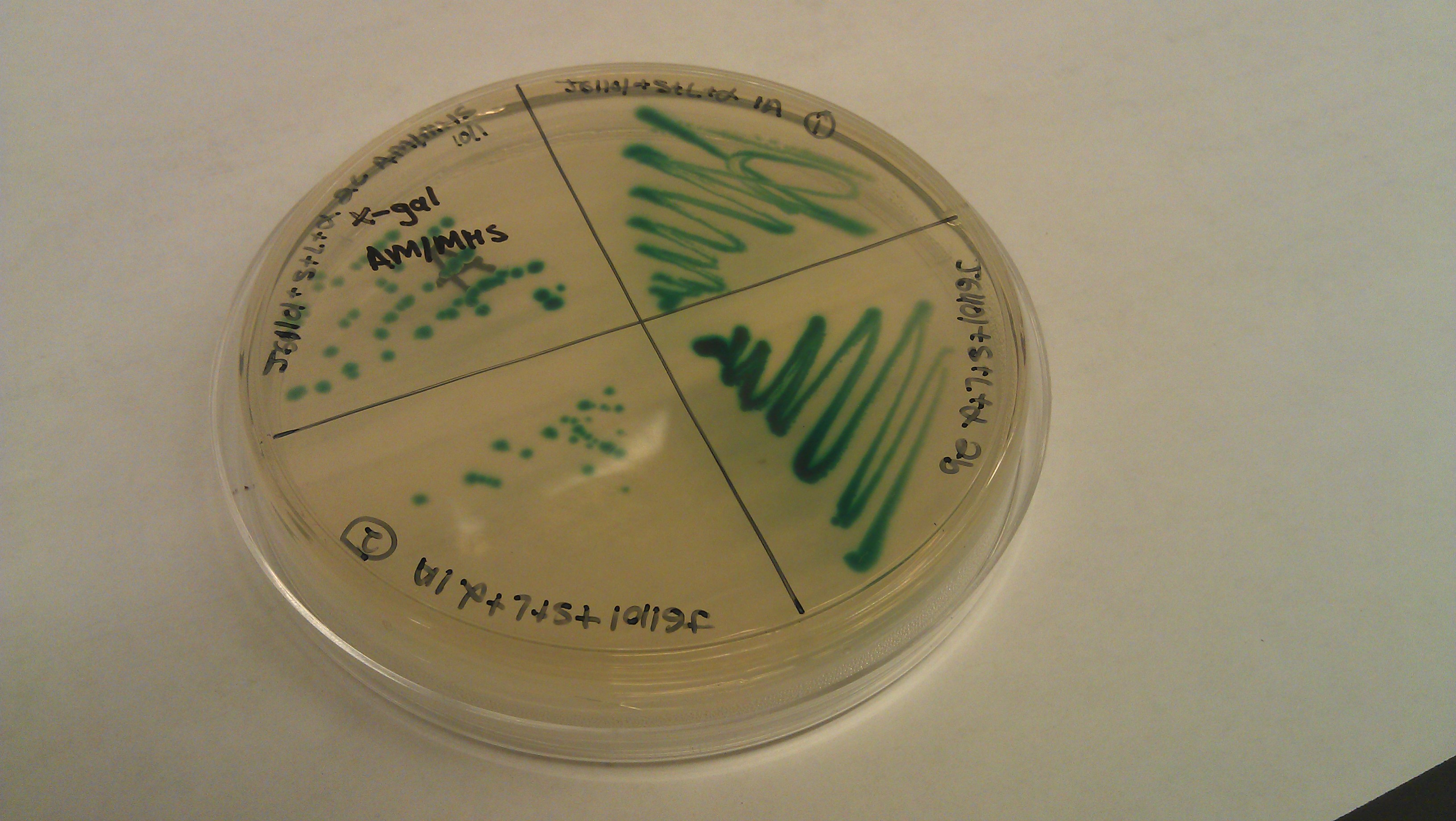
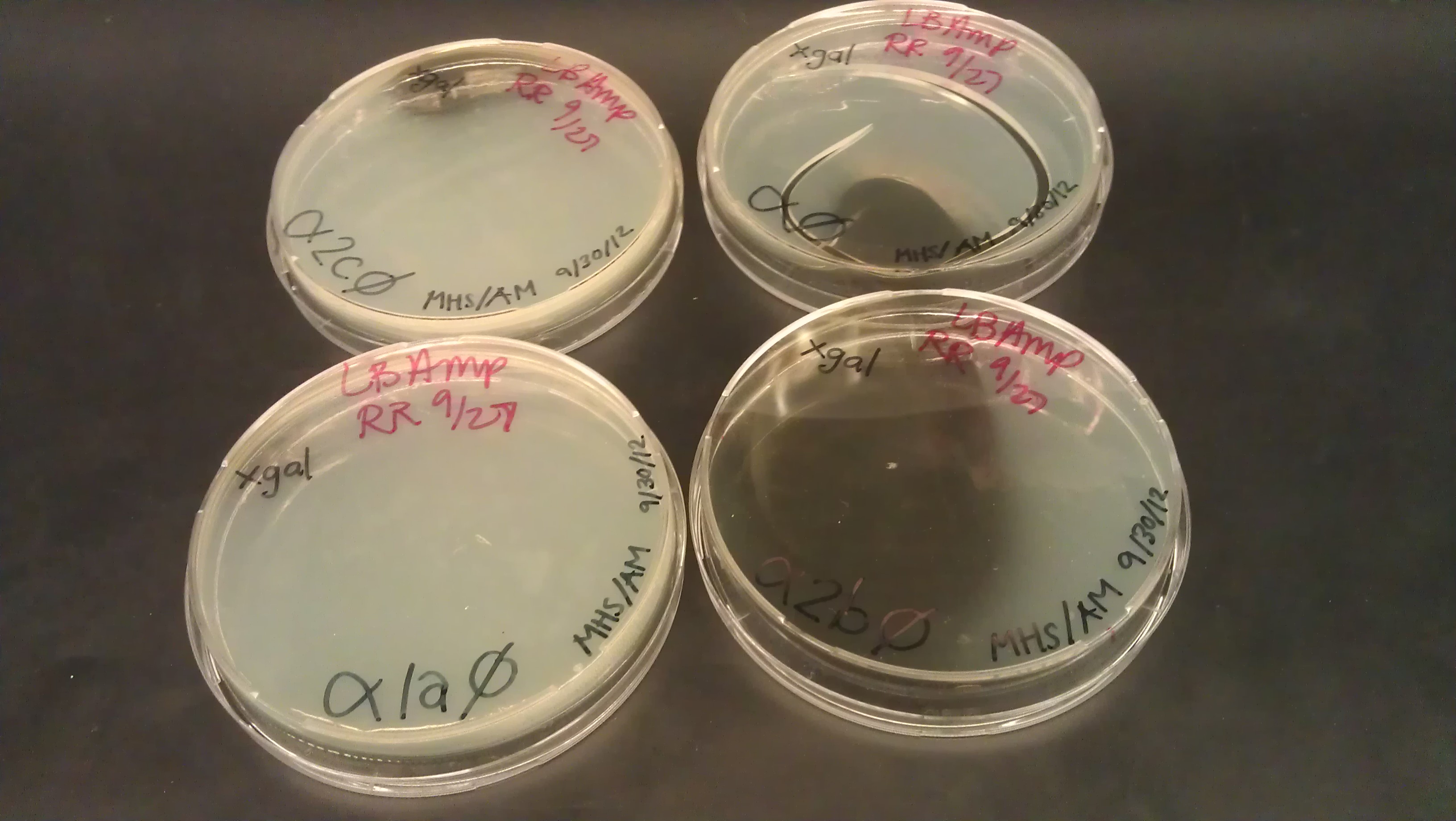
- J61100 and J61101 constitutive promoters were extracted from the distribution plates and an additional promoter, pLux-Lac+RBS, were used to prove that the probe functions as desired.
- Following assemblies were set up:
- Plated on 50ul xgal LB amp Plates
- J61100+ α
- J61100+ strep+L+α (1a)
- J61100+ strep+L+α (2c)
- J61100+ strep +L +α(2b)
- J61101+ α
- J61101+strep+L+α(1a)
- J61101+strep+L+α (2c)
- J61101+strep+L+ α (2b)
- plux-lac1+rbs+α
- plux-lac1+rbs+strep+L+α(1a)
- plux-lac1+rbs+strep+L+α(2c)
- plux-lac1+rbs+strep+L+α(2b)
- plux-lac2+rbs+α
- plux-lac2+rbs+strep+L+α (1a)
- plux-lac2+rbs+strep+L+α (2c)
- plux-lac2+rbs+strep+L+α (2b)
- Following were plated on 50ul X-Gal and 25ul IPTG LB AMP plates (no growth expected)
- plux-lac1+rbs+α
- plux-lac2+rbs+strep+L+α (1a)
- plux-lac2+rbs+strep+L+α (2c)
- plux-lac2+rbs+strep+L+α (2b)
- Plate pictures:
Topo
- Miniprepped GFPT1, GFPT2, Topo, and Topo D168A (Topo cultures separated into 3mL and 1mL minipreps)
- Nanodropped miniprepped DNA:
- GFPT1 I - 275.29 ng/uL
- GFPT1 II - 101.08 ng/uL
- GFPT2 I - 222.11 ng/uL
- GFPT2 II - 230.52 ng/uL
- Topo I - 117.97 ng/uL
- Topo II - 70.28 ng/uL
- Topo D168A I - 69.47 ng/uL
- Topo D168A II - 51.87 ng/uL
- HIS purified crude lysates from 9/27/12 (Topo, Topo D168A, Topo + GFPT1, Topo + GFPT2, all IPTG induced)
- Digested Topo I plasmid (pet29a) with E and X
- Ran a gel with Hyperladder I, 1-2 1-3 4-5 and 6-7 PCR products, pet29a digestion, and pSB1C3 digestion
- Confirmed that PCR made amplicons
- Excised bands for digested pet29a and pSB1C3 plasmids
- Gel extracted 4 gel fragments (2 wells per sample: digested pSB1C3 plasmid, digested pet29a plasmid)
- Nanodropped gel extractions:
- Digested pSB1C3 - 25.54 ng/uL
- Digested pet29a - 22.95 ng/uL
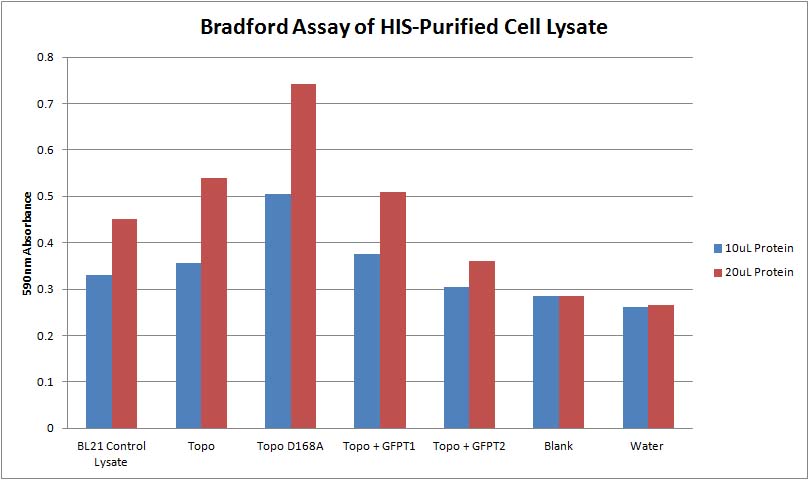
- Set up a bradford assay of topo, topo D168A, topo + G1, topo + G2 (5uL protein, 10uL protein, 20uL protein + 200uL reagent)
- Used ~100uL aliquot of BL21 competent glycerol stock to seed 10mL of LB medium (no antibiotic), stored at 37C
- Ran 1% agarose gel with samples: A1, B1, A2, B2 and Hyperladder I
- Did bug buster protocol to lyse BL21 control culture (used lysonase)
- Ligated digested pet29a with pet29 top/bot annealed oligos
- Transformed ligation using invitrogen DH5alpha transformation protocol
- Prepared LB kanamycin plates
- plated transformed ligations on prewarmed kanamycin plates
September 30
Topo
- All assemblies were successful and went as planned. Blue colonies were picked to generate liquid cultures and streak plates to have a better visual result.
- Pet29a plates did not grow
- Kinase treated pet29a oligos
- Annealed kinase treated pet29a oligos
- Ligated digested pet29a (from gel extraction) with kinase treated oligos
- Transformed ligations into DH5alpha, used topo plasmid as a positive control
- Plated transformations on kanamycin plates and stored overnight at 37C
- HIS purified BL21 control crude lysate
- Set up a bradford assay with:
- uninduced & induced topo protein extractions from 9/19
- uninduced & induced topo D168A protein extractions from 9/19
- BL21 control lysate
- Topo, Topo D168A, Topo + G1, Topo + G2 from 9/27
- All samples prepared (10uL protein, 20uL protein + 200uL reagent)
- Miniprepped GFPT1 1,2,3 and GFPT2 (17,18,26) (~600uL of each) (1mL liquid cultures made from colonies on the chloramphenicol plates of GFPT1 and GFPT2 ligated into the shipping vector)
- Nanodropped:
- GFPT1-1 - 38.5 ng/uL
- GFPT1-2 - 77.6 ng/uL
- GFPT1-3 - 74.8 ng/uL
- GFPT2-17 - 61.6 ng/uL
- GFPT2-18 - 65.9 ng/uL
- GFPT2-26 - 64.2 ng/uL
- Ran a 1% agarose gel with 1-1, 1-2, 1-3, 2-17, 2-18, 2-26 plasmid miniprep samples and Hyperladder I
- Prepared a 1:2 dilution of GFPT2 plasmid from 9/27 miniprep
- Treated 5uL of diluted plasmid with 5uL of water, BL21 protein, topo protein, topo D168A protein
- Incubated 30 minutes at 37C
- Ran a 1% agarose gel with protein treated target plasmid samples and Hyperladder I
- Digested GFPT2 plasmid with X (let run at 37C for 30 minutes)
- Used DNA clean up kit on digested GFPT2
- Nanodropped digested GFPT2:
- GFPT2(X) - 39.85 ng/uL
- Prepared DNA seq samples using VF2 and VR
- Sample# - PrimerPair - DNA sample (sample 1, GFPT2 uncut + VF2; sample 2, GFPT2 uncut + VR)
- 1/2 - FWD/REV - uncut GFPT2 plasmid
- 3/4 - FWD/REV - cut GFPT2 plamid
- 5/6 - FWD/REV - 2:1 uncut:cut GFPT2 plasmid mixture
- 7/8 - FWD/REV - 1:1 uncut:cut GFPT2 plasmid mixture
- 9/10 - FWD/REV - 1:2 uncut:cut GFPT2 plasmid mixture
- 11/12 - FWD/REV - 1-2 miniprep sample from 9/19 double transformations
- 13/14 - FWD/REV - 1-2I
- 15/16 - FWD/REV - 1-3
- 17/18 - FWD/REV - 1-3I
- 19/20 - FWD/REV - 2-1
- 21/22 - FWD/REV - 2-1I
- 23/24 - FWD/REV - 2-2
- 25/26 - FWD/REV - 2-2I
- 27/28 - FWD/REV - 2-3
- 29/30 - FWD/REV - 2-3I
October 1
Strep
- Strep samples were minipreped and digested. The samples were inserted into the shipping vector.
- Minipreps of:
- J61011 + S + L + ALPHA 2B
- J61100 + S + L + ALPHA 2B
- J61100 + S + L + ALPHA 2C
- PLUX2 + RBS + ALPHA 1A
- PLUX2 + RBS + ALPHA 1A (2)
- PLUX + S + L + ALPHA 2C
- J61101 + S + L + ALPHA 1A
- J61101 + S + L + ALPHA 1A (2)
- PLUX2 + RBS + S + L + ALPHA 2B
- PLUX2 + RBS + S + L + ALPHA 2B (2)
- J61101 + S + L + ALPHA 2C
- PLUX + S + L + ALPHA 2B
- Ran Twice when finding concentrations:
- J61101 + S + L + ALPHA 1A
- J61101 + S + L + ALPHA 2B
- Restricted all the above and:
- Strep + Linker + OMEGA
- Strep + Linker-4 + OMEGA
- with:
- X+P
- Gel confirmed
- Ligated into pSB1C3 shipping vector and transformed into BL21(DE3) cells.
- Prepared all 14 above samples for sequencing.
October 2
Strep
- Transformation failed and DNA was turned in for sequencing.
Topo
- Prepared PCR tubes with 10uL Topo1 4 (25ng/uL topo d168a in the shipping vector), 10uL GFPT2-26 (25ng/uL GFPT2 in the shipping vector), and 10uL GFPT1-3 (25ng/uL GFPT1 in the shipping vector)
- Labelled the tubes K891234, K891999, K891000 respectively and shipped overnight to iGEM Headquarters
- Topo D168A treated DNA samples, incubated for 10 minutes at 37C:
- Omega fragment PCR amplicon
- Topo coding sequence PCR
- GFPT1 VF2/VR PCR amplicon
- Ran a 1% agarose gel containing untreated omega PCR, untreated topo PCR, untreated GFPT1 PCR, treated omega PCR, treated topo PCR, treated GFPT1 PCR, and Hyperladder I
October 3
Topo
- Protein treated GFPT2 plasmid, incubated for 10 minutes at 37C:
- BL21 HIS-purified Control Lysate
- Topo
- Topo D168A
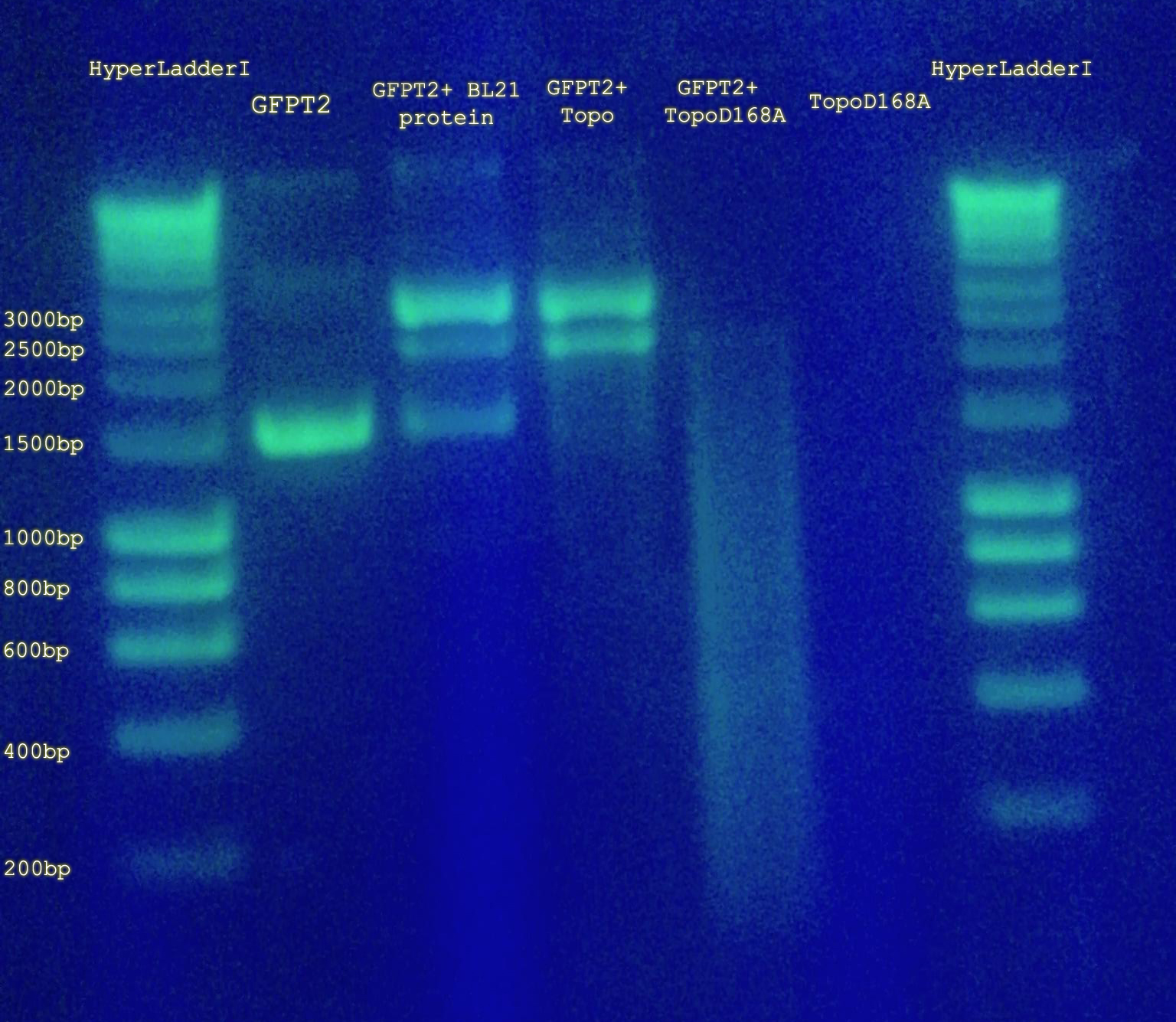
- Ran a 1% agarose gel containing GFPT1, GFPT1 + BL21 protein, GFPT1 + Topo, GFPT1 + Topo D168A, Topo D168A (no DNA), and Hyperladder I
October 18
Magainin
- Overlapping oligo assembly of Magainin + Linker + His-tag
- Ligated assembly into GFP vector BBa_I13522 for green-white screen
- Transformed into DH5a cells and incubated overnight with negative control
October 19
Magainin
- Picked a single white colony from overnight plate and made liquid culture.
Topo
- Biobricked template: pet29a adapter F and R - cut with X&E, ligate into open pet vector
- PSV template: 1 - omega R + SP and 1-Omega F+TSE
- Topo in pet29a template: Topo R omega His + TSE and T7 forward adding X
- Topo in pet29a template: Topo R alpha His+S and T7 forward adding X
- PSV template: alpha-4 F+X and alpha-4 R+SP
- PSV template: new R alpha SC+SP and 6:F beta-gal-alpha X
- 5:R Beta-gal-omega SP & 4F: beta-gal-omega TSE --> PSV template
- Cut PCR products & ligate
- 3 SpeI + 4Xbai, 5Xbai ligate --> cut X&P, drop into appropriate vector
- 2 TSE + 1 TSE, 6 TSE ligate --> cut X&P, drop into appropriate vector
- Cut PCR products & ligate
- Made liquid cultures off of plates:
- T7 (1:15N)
- PSV
- magainin (in psb1A2)
October 20
Magainin
- Miniprepped Magainin + Linker + His-tag +BBa_I13522 (274 ng/uL)
- Cut with S+P and ligated to alpha-4 fragment cut with X+P.
- Cut with S+P and ligated to omega fragment cut with X+P.
- Transformed both ligations into DH5a cells with negative control
Topo
- Miniprepped: T7, PSV, magainin
- gel visualization of PRC products from 10/19/12 (all of them worked)
- PCR clean-up of PCR products
- Digestion & Ligation of PCR Products (pet 29 adapter, E&X)
- Digestion of TSE fragments (15 uL PCR product, 3uL NEB4 buffer, 1 uL TSE, 11 uL H2O)
- Digestion with S&X
- ligated and ran on a gel
- restarted PCRs (Topo+Alpha, Alpha, Alpha-4, Omega)
- 1 hour anneal times for omega topo alpha
- 2 hour anneal times for omega topo alpha-4
- 27 ccles 55 anneal temp
- mixed matching PCR products
- PCR cleaned (kit) PCR products; eluted in 30 uL
- Ligated EX digest pet adapter to EX open isolated pet plasmid backbone
- Transformed into DH5alpha
- outgrowth in SOC
- plated on kanamycin plate; stored @ 37C
- Digested 4 PCR products: Beta-gal fragments; Topo w/ S
- Digested T7 plasmid S&P
- PCR cleaned T7, and 3 ligates eluted in 15 uL
- Digested all 3 ligations with X & P for 30 min @ 37C
- Ran on a gel hyperladder 1
- Excised 1100, 3000, 2000 bp bands
- Gel extracted eluted w/ 8 uL
- Ligated T7: let run @ room temp for 30 min; transformed into DH5alpha, seeded into amp plates; outgrowth in SOC
October 21
Magainin
- Picked single colonies of both Magainin + Linker + His-tag + alpha-4 and Magainin + Linker + His-tag + omega and made liquid cultures.
Topo
- Colony PCR on 6 colonies from each topo fusion plate
- Made master mix, prepared 10 uL PCR reactions for each colony
- Picked half of each colony and swirled tip in PCR mix
- Used beta-gal primers for each respective part: alpha, alpha-4, omega (25 cycles annealing temp)
- Digest ~5uL (100ng/uL) T7 promoter at 37C for 45 min
- PCR cleaned digestion
- Transformed into DH5alpha (followed Haynes 101 procedure)
Ran PCR product on gel w/ hyperladderI
- No bands anywhere (thick primers, smear in all wells, thicker in first 7 samples)
- made liquid cultures of
- Amp: alpha, omega
- Kanamycin: pet+adapter
- stored alpha, omega, & alpha-4 in 4C
October 22
- Miniprepped alpha (1) and omega (1) and (2)
- Restricted 4uL each of topo alhp and topo omega with 1 uL EcoRI
- Ran on gel
- Ran 5 uL of unrestricted plasmids on a gel topo alpha, omega
- Colony PCR'd alpha-4 and T7 with VF2 and VR
- Ligated T7 promoter from 10/21/12 to topo + alpha-4
- Transformed into DH5alpha
- Outgrowth in 50C for an hour at 37C
- Plated on amp plate
October 23
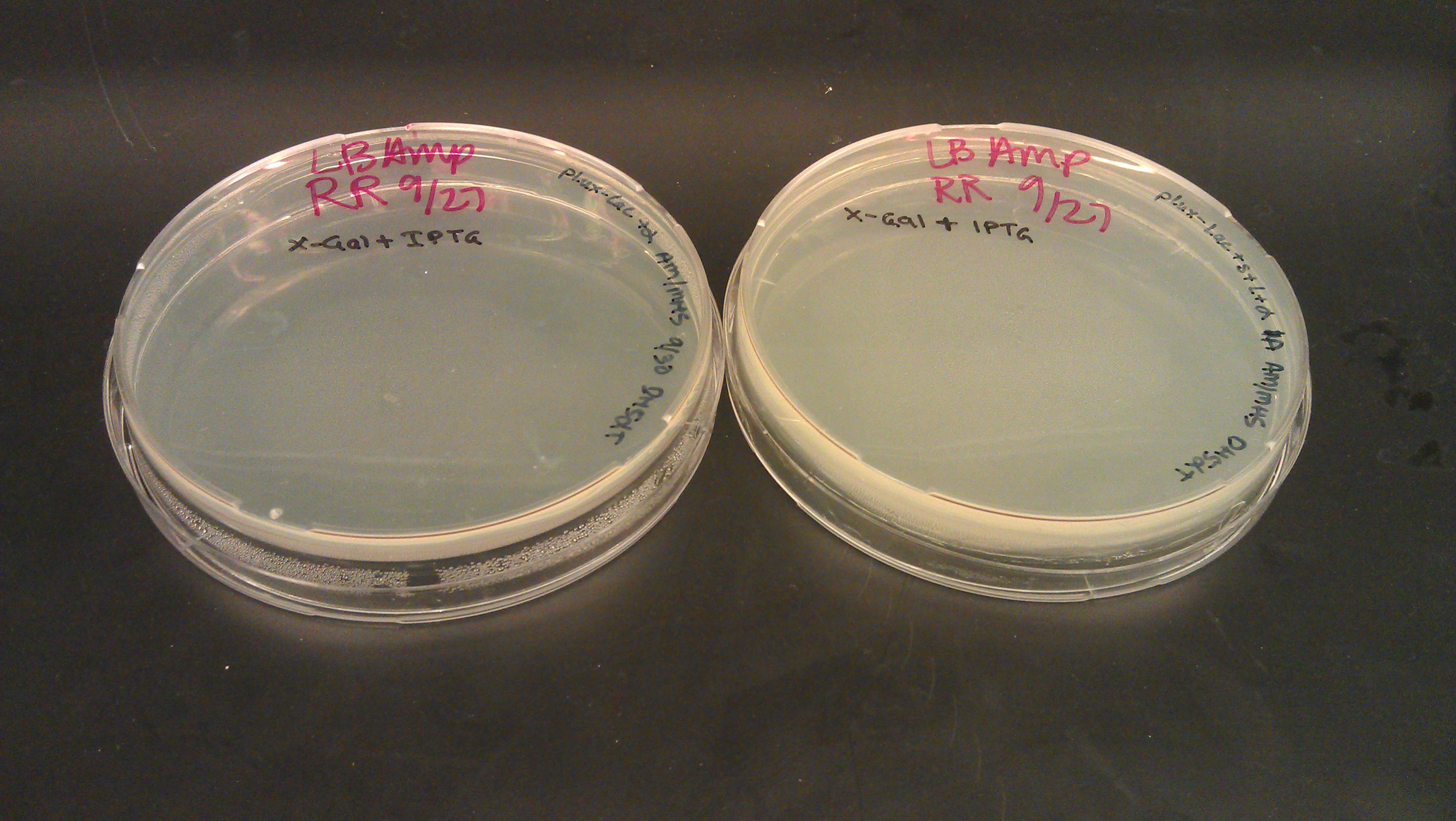
Water Sample Testing
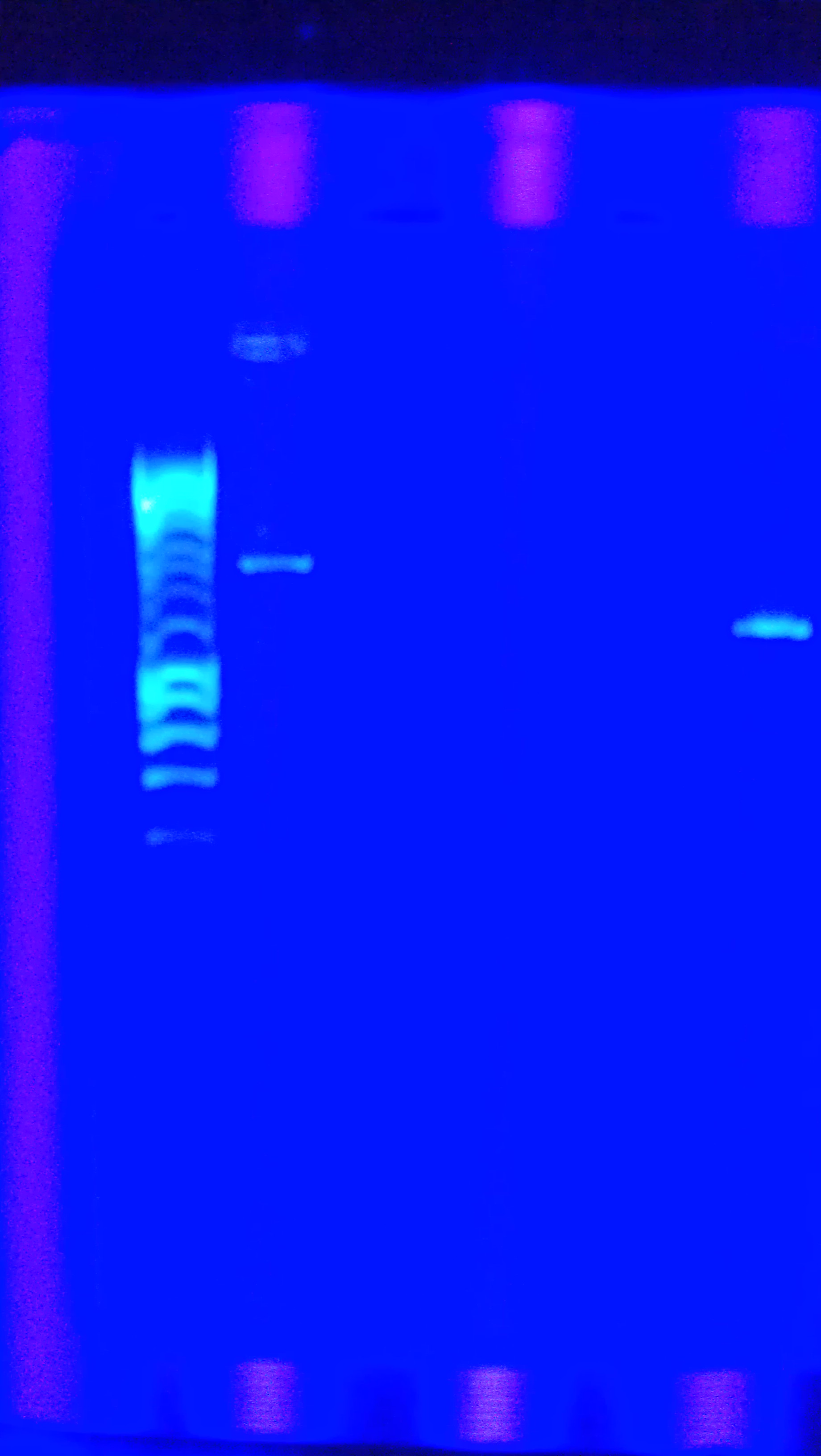
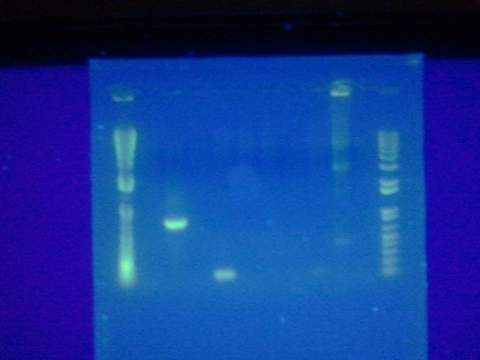
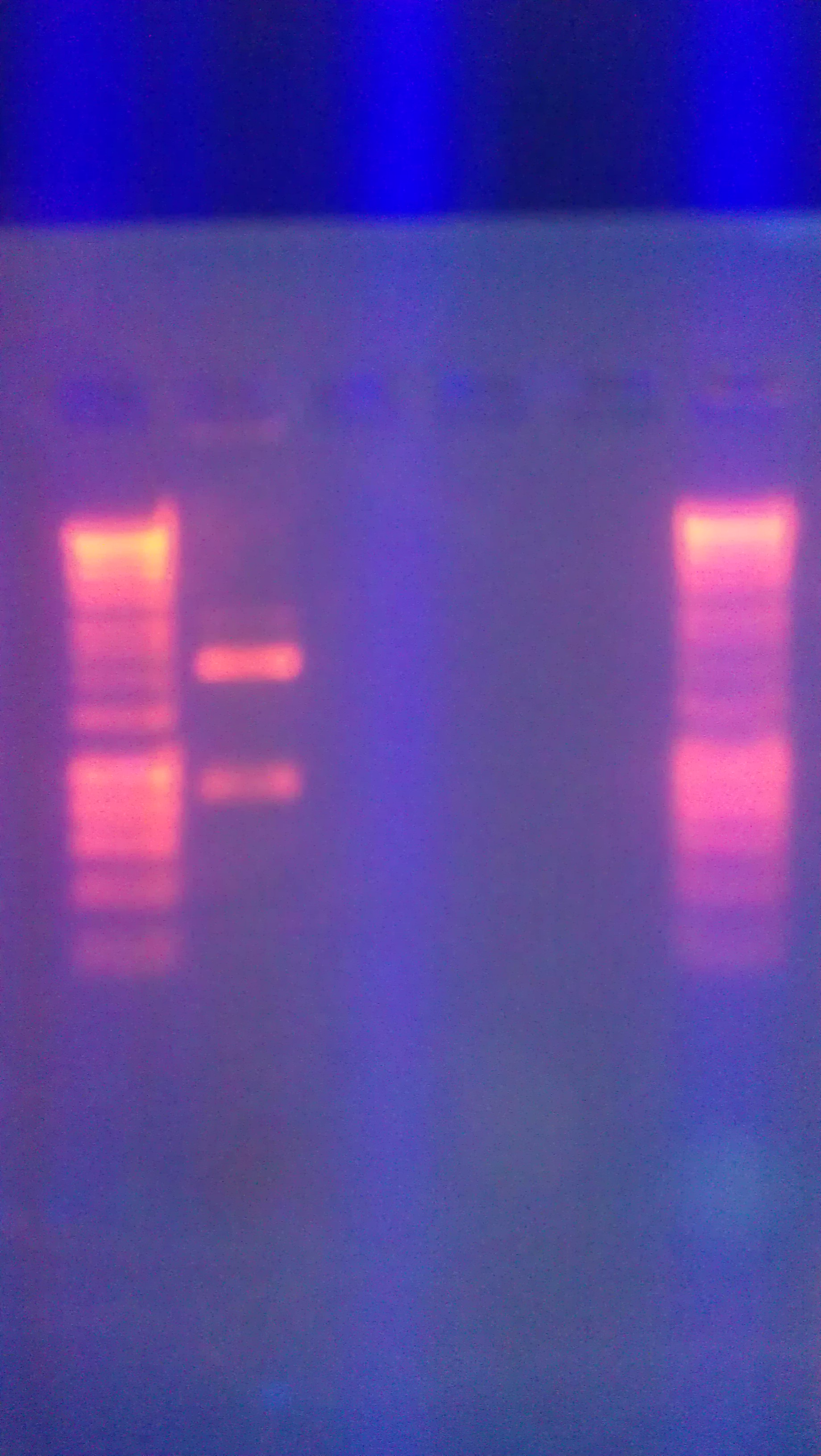
- Figure 1: Left to right, water sample from Artisan Courtyard fountain (ASU campus), water sample from Tempe Town Lake and Nuclease free water. The following picture was taken 10 minutes after X-gal, PSV cell lysate (contains full LacZ) and DTT were added to the solution. The following test samples were under buffering conditions. All three samples changed colors but the Tempe town lake water sample reflected a color change 2 minutes after the addition of the reagents.
- Figure 2: Left to right, Nuclease free water sample, Tempe Town Lake water sample and Artisan Courtyard fountain (ASU campus) water sample. The following picture was taken 10 minutes after X-gal, PUC 19 cell lysate (contains split LacZ fragements) and DTT were added to the solution. The following test samples were under buffering conditions. Faint color change was observed in all three samples.
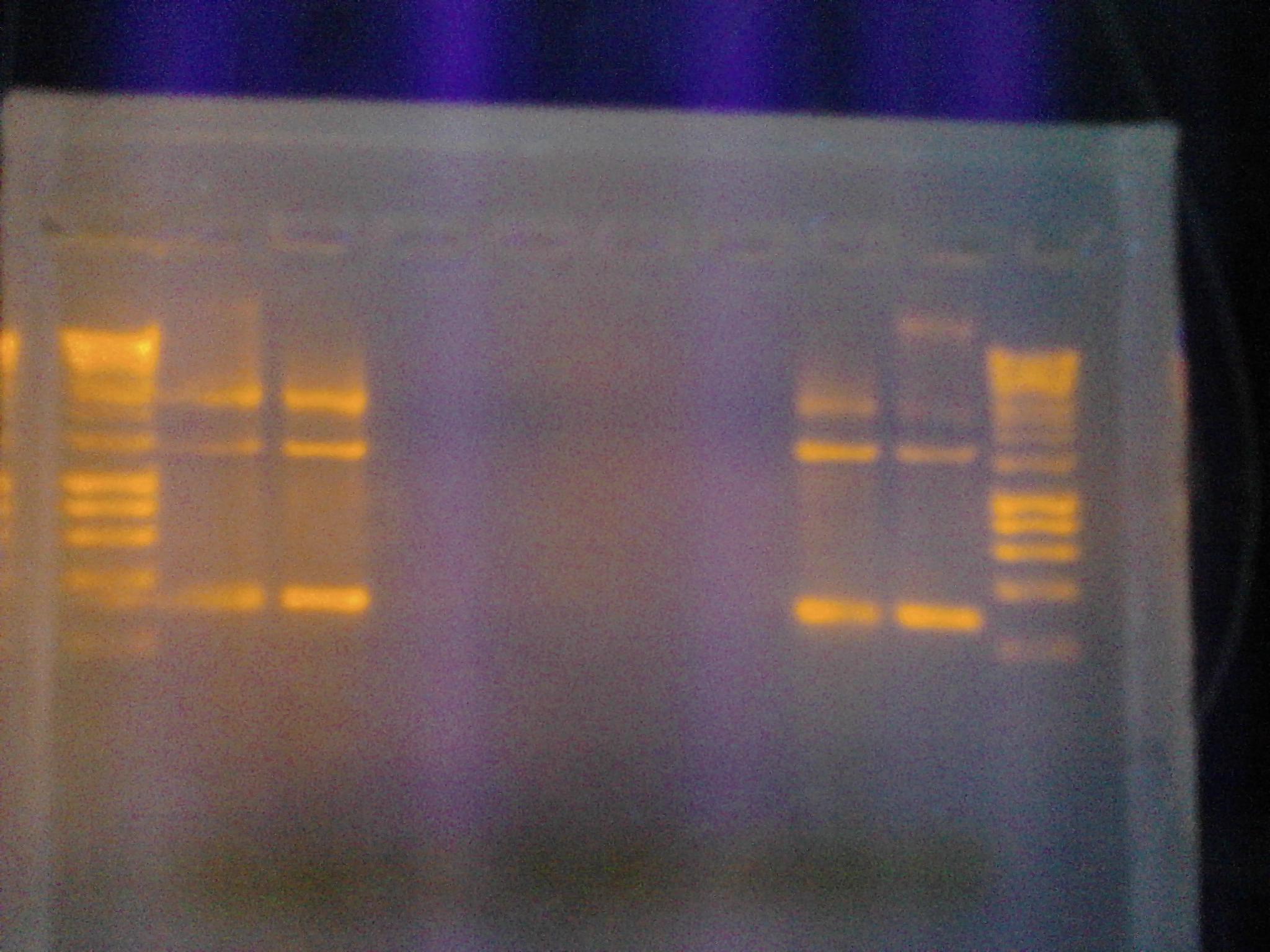
- Figure 3: Control Samples. Left to right, water sample from Artisan Courtyard fountain (ASU campus), water sample from Tempe Town Lake and Nuclease free water. The following picture was taken 10 minutes after X-gal, NEB10 B cell lysate (contains omega fragment of LacZ) and DTT were added to the solution. The following test samples were under buffering conditions. No color change was observed.
- Figure 4: Left to right, water sample from Artisan Courtyard fountain (ASU campus), Nuclease free water and water sample from Tempe Town Lake. The following picture was taken 10 minutes after X-gal, commercial beta gal enzyme and DTT were added to the solution. The following test samples were under buffering conditions. No color change was observed.
NEED PICTURE
- Figure 5: Left to right, water sample from Tempe Town Lake, Nuclease free water and water sample from Artisan Courtyard fountain (ASU campus). The following picture was taken 10 minutes after X-gal and commercial beta gal enzyme were added to the solution. The following test samples were not under buffering conditions. No color change was observed.
NEED PICTURE
- Figure 6: Left to right, water sample from Tempe Town Lake, Nuclease free water and water sample from Artisan Courtyard fountain (ASU campus).
NEED PICTURE
- Figure 7: Figure 1, observed after 10 minutes (Left to right, water sample from Tempe Town Lake, water sample from Artisan Courtyard fountain (ASU campus) and Nuclease free water). Samples are attaining a darker blue shade.
NEED PICTURE
- Figure 8: Figure 2, observed after 10 minutes. Samples still only have faint blue color.
NEED PICTURE
- Figure 9: Figure 3, observed after 10 minutes. No color change.
NEED PICTURE
- Figure 10: Figure 4, observed after 10 minutes. No color change.
NEED PICTURE
- Figure 11: Figure 5, observed after 10 minutes. No color change.
NEED PICTURE
- Figure 12: Figure 7, observed after 10 minutes. All the samples turned blue.
NEED PICTURE
- Figure 13: Figure 8, observed after 10 minutes. Water samples from the Lake and Nuclease free water had a deeper blue color compared to the sample from Artesian Courtyard fountain.
NEED PICTURE
- Figure 14: Figure 9, observed after 10 minutes. No color chance as expected.
NEED PICTURE
- Figure 15: Figure 10, observed after 10 minutes. No color change.
NEED PICTURE
- Figure 16: Figure 11, observed after 10 minutes. No color change.
NEED PICTURE
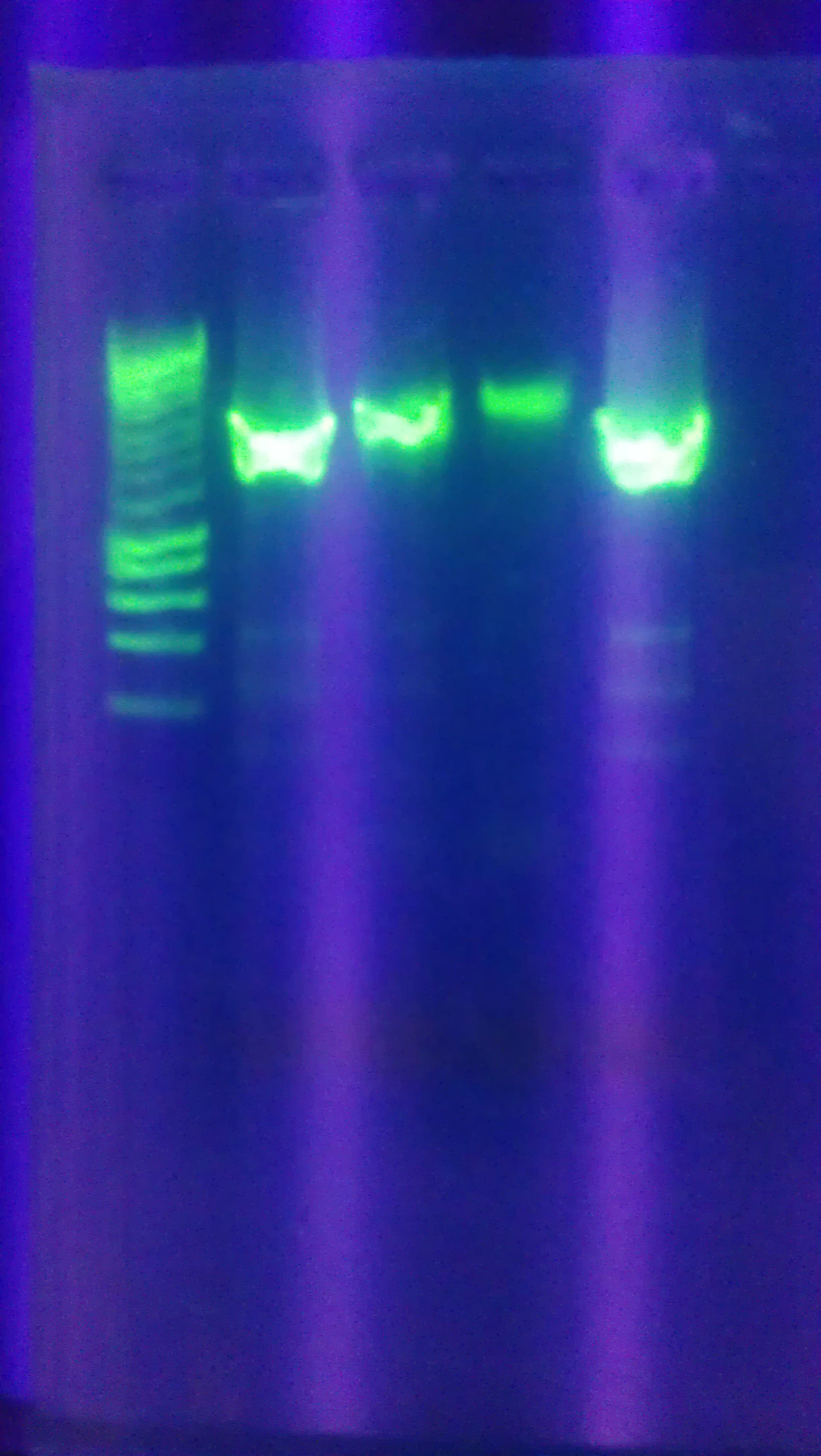
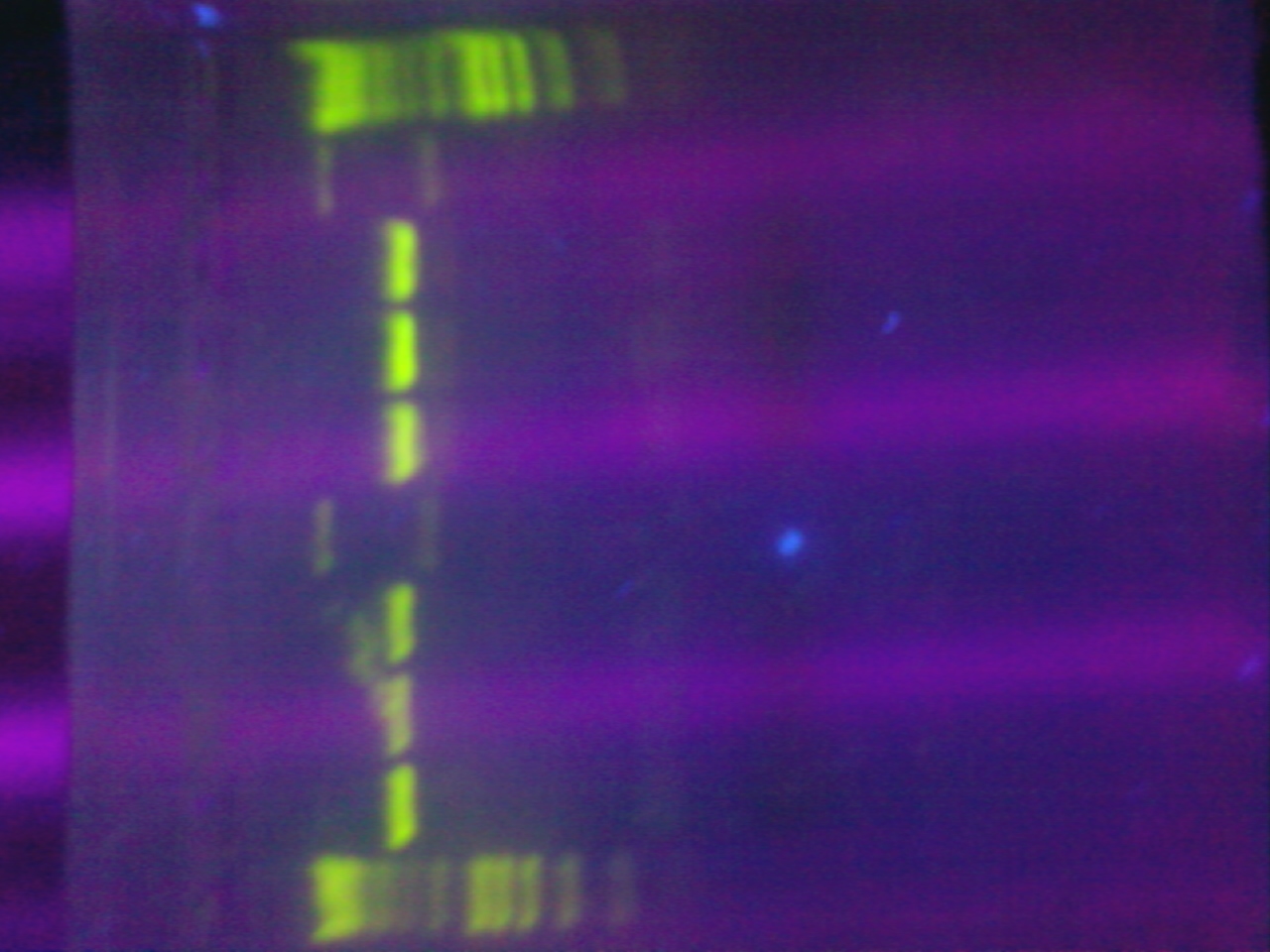
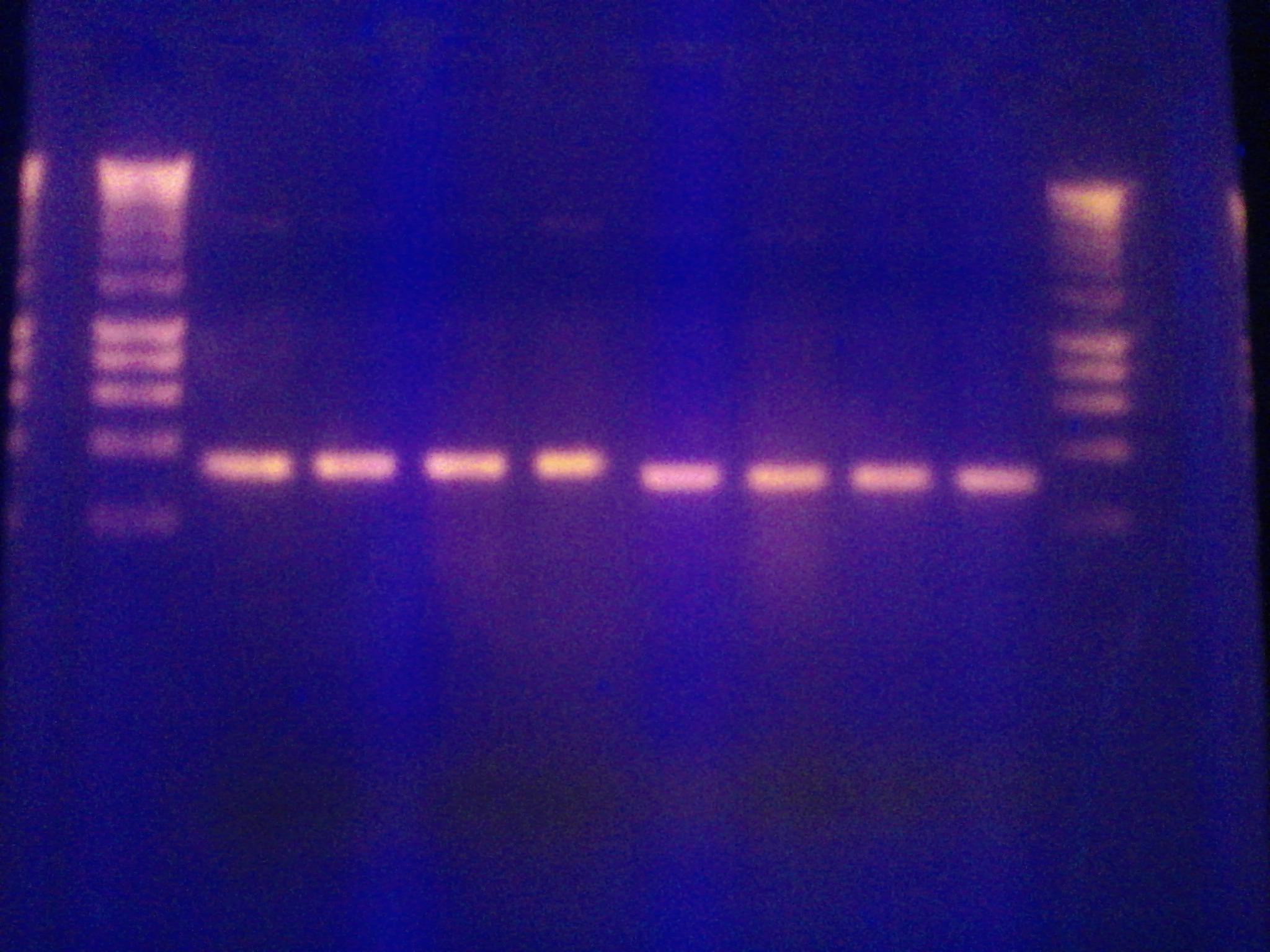
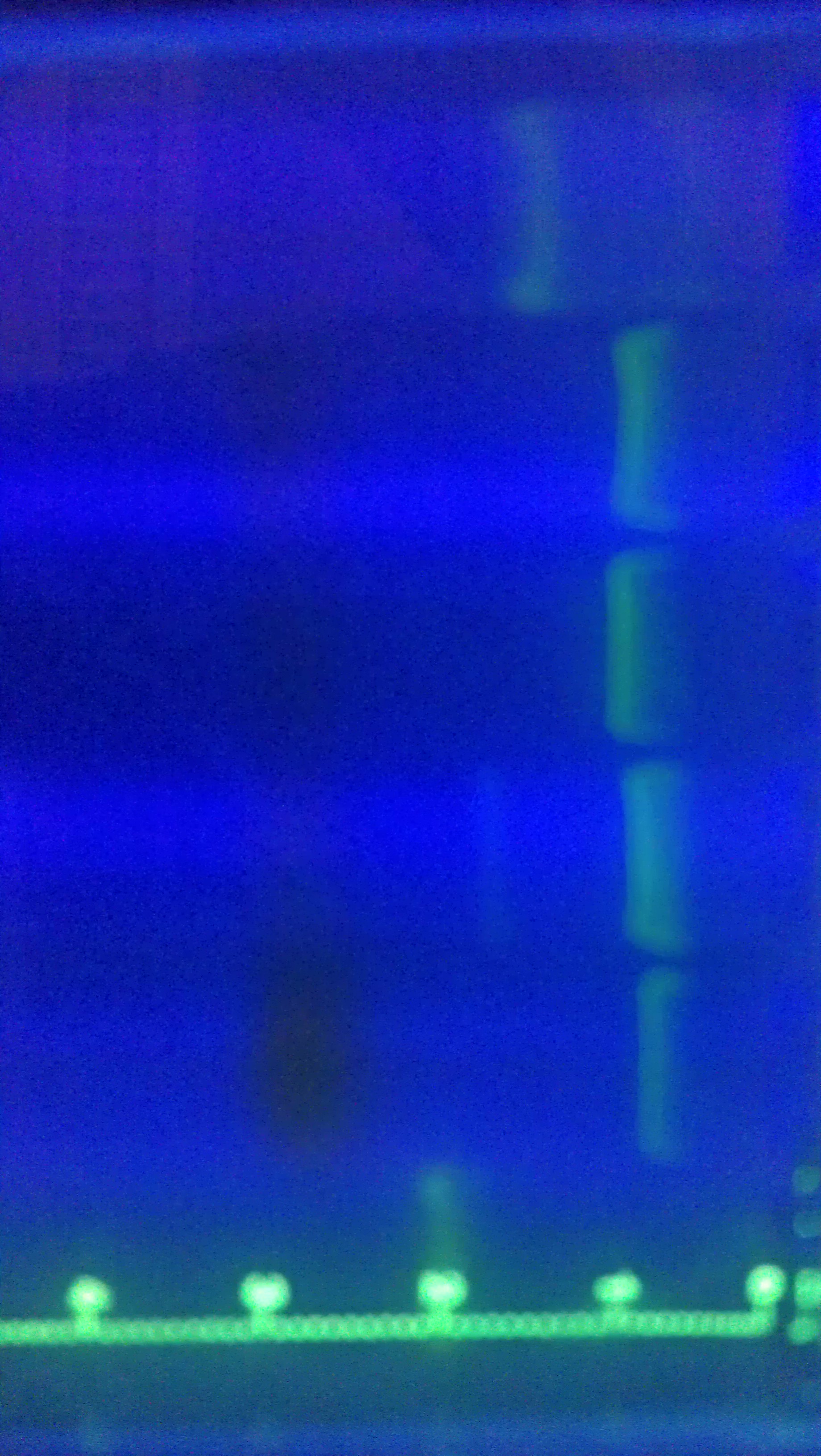
- Ran colony PCR of 8 colonies from each of the three transformed plates from 10/23 (alpha, alpha 4,omega)
- Used VF2 and VR biobrick sequencing primers
- 55C annealing temperature
- 3.5 min elongation time
- Used VF2 and VR biobrick sequencing primers
- Visualized the PCR product on a gel.
October 24
- PCR amplification of:
- Topo adding XbaI site in front, SpeI site behind
- Alpha 4 adding XbaI site in front, SNP biobrick suffix behind
- Alpha adding XbaI site in front, SNP biobrick suffix behind
- Omega adding XbaI site in front, SNP biobrick suffix behind
- PCR mutation of:
- Topoisomerase coding sequence (Bba_K891234) to remove EcoRI site at 896th nucleotide
- Omega coding sequence to remove internal EcoRI site.
- Made liquid cultures of Topo+GFPT1 and Topo+GFPT2
October 25
- SOEing PCR for mutation of Topo coding sequence and Omega coding sequence
- 25uL Syzygy PCR master mix
- 12.5uL PCR front mutational half
- 12.5uL PCR reverse mutational half
 "
"