Team:Chalmers-Gothenburg/Theory
From 2012.igem.org
| Line 1: | Line 1: | ||
{{Team:Chalmers-Gothenburg/Template2}} | {{Team:Chalmers-Gothenburg/Template2}} | ||
| - | <div align="justify" style="width:auto; margin-left:0px; margin-right: | + | <div align="justify" style="width:auto; margin-left:0px; margin-right:0px;"> |
==Introduction== | ==Introduction== | ||
In this section we present some information needed to understand the design of our biosensor. | In this section we present some information needed to understand the design of our biosensor. | ||
Revision as of 11:24, 9 July 2012
Contents |
Introduction
In this section we present some information needed to understand the design of our biosensor. Firstly, a short introduction to G protein-coupled receptors will be given. Thereafter, the hCG hormone, its associated LH/CG receptor and their roles during pregnancy will be discussed. The construction of our hCG biosensor will require the assembly of several biological components in yeast, such as a functional human GPCR, a modified pheromone signaling pathway, and reporter genes encoding indigo-synthesizing enzymes. Hence, in this section, we will also present needed background information and previous approaches within these areas.
Human chorionic gonadotropin hormone
The hormone of interest in detecting pregnancy is human chorionic gonadotropin hormone (hCG), a member of the glycoprotein family [1]. hCG binds to luteinizing hormone (LH) receptor localized on the yellow body that will after binding of hCG maintain progesterone production. The maintained level of progesterone stops menstrual bleeding and prepares the uterus lining for the implementation of the fertilized egg [2]. The term hCG actually refers to five independent molecules, all with various structure and biological function. The two dominating forms are regular hCG and hyperglycosylated hCG (H-hCG) but only regular hCG acts on the LH receptor. Therefore our biosensor will only detect regular hCG [3].
Luteinizing hormone receptor
The LH/CG receptor belongs to the family of G proteins-coupled receptors characterized by an extracellular domain, seven transmembrane domains and coupling to a G protein [4]. Several studies has investigated the hCG and LH/CG receptor interactons and it has showed that parts all over the receptor is important for ligand binding and signal transduction [5]. The extracellular domains of the LH/CG receptor also contains six potential sites for glycosylation and some of the glycosylation sites may affect the intracellular transport, membrane insertion or recycling of the receptor [6].
Yeast pheromone pathway
S.cerevisiae stably exists as either a haploid or a diploid. In both modes of existence, it reproduces asexually through a process called budding. This is an asymmetrical dividing process in which a mother cell gives rise to a smaller daughter cell [7]. Diploid yeast cells are stable but under stress conditions such as absence of nutrition, they can form haploid spores. In the haploid mode, yeast has two mating types, a and α called MATa and MATα respectively. The haploid yeast cells are capable to mate with the opposite mating type which results in the formation of a diploid zygote [8].
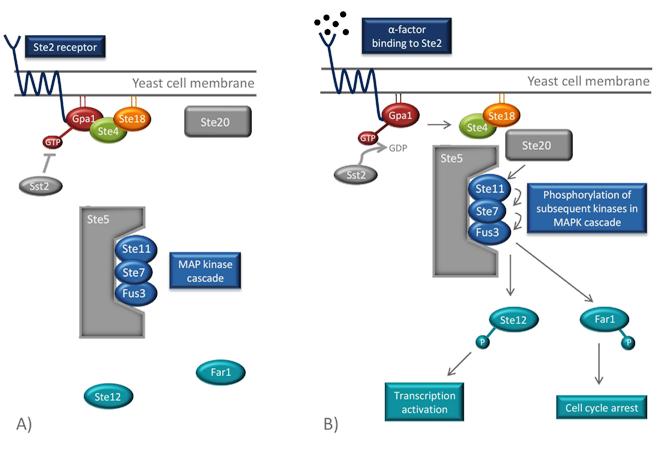
Mating between haploid cells is mediated by diffusible molecules called pheromones. The mating types secrete different pheromones (a- and α-factors) and respond to the mating factor of the opposite mating type. The response occurs when the pheromones bind to a G protein-coupled receptor in the yeast cell membrane. The two mating types have different GPCRs (Ste2 and Ste3) each binding to the corresponding pheromone. The pheromones activate the GPCR which in turn creates an intracellular response. This intracellular signal transduction pathway in yeast is one the best understood signaling pathways in eukaryotes [9] and will also be utilized in the hCG biosensor. The whole pathway is illustrated in the Figure below.
Signal transduction
The GPCR of the MATa mating type is Ste2. Its associated G protein at the inside of the cell membrane consists of three subunits, Gpa1, Ste4 and Ste18, which correspond to mammalian Gα, Gβ and Gγ [9]. When α-factors from the MATα cell bind to the Ste2 receptor of the MATa-cell, a conformational change of the GPCR occurs. This activates the α subunit of the G protein, the Gpa1, by exchanging its bound GDP for GTP. This leads to the dissociation of the Ste4-Ste18 complex, which in turn binds the effector proteins Ste5 and Ste20, through Ste4. Ste20 is a protein kinase and Ste5 is a scaffold protein that binds and organizes different kinases of the mitogen-activated protein kinase (MAPK) cascade. When the Ste4-Ste18 complex binds to both Ste5 and Ste20, the two proteins are moved towards each other. This enables Ste20 to phosphorylate the first kinase in the MAPK cascade, Ste11, which is bound to Ste5. Consequently, subsequent kinases (Ste7 and Fus3) of the cascade are phosphorylated, which ultimately results in the phosphorylation and activation Far1 and Ste12 [9].
Indigo
Bio-indigo can be synthesized from the amino acid tryptophan in a few simple steps, which are outlined in the picture below. The first reaction is the conversion of tryptophan to indole, catalyzed by tryptophanase. The next part of the pathway is catalyzed by a mono- or dioxygenase and transforms indole to indoxyl. Indoxyl is then converted to indigo by several spontaneous reactions in the presence of oxygen [10], [11]. In order to produce bio-indigo in S. cerevisiae, genes encoding tryptophanase and a flavin-containing monooxygenase are inserted.
 "
"