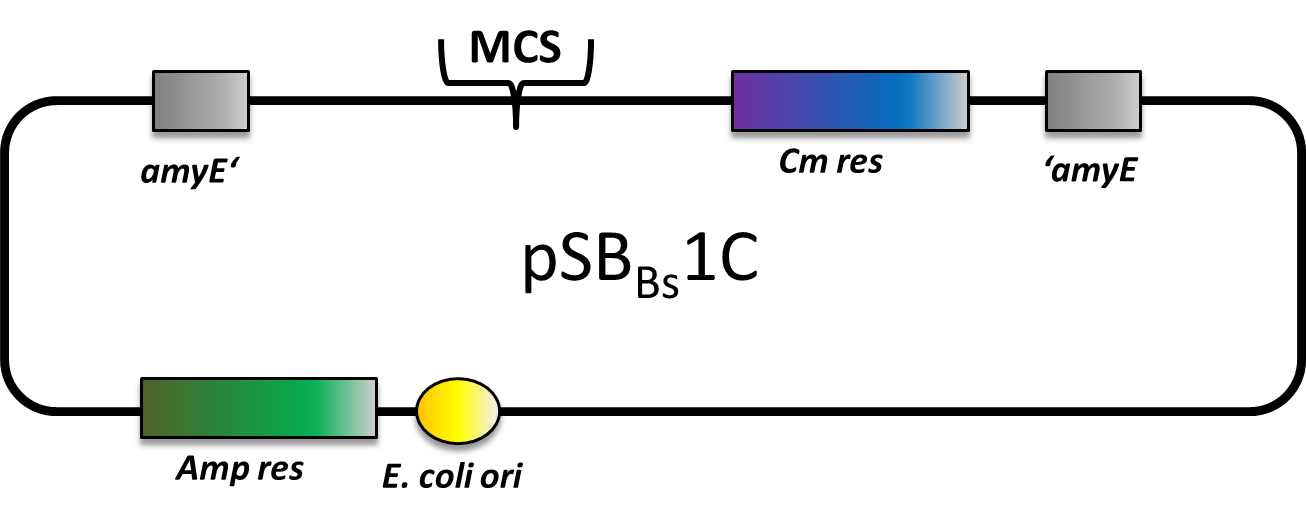Team:LMU-Munich/Bacillus BioBricks
From 2012.igem.org
Franzi.Duerr (Talk | contribs) |
(Vector Box) |
||
| Line 53: | Line 53: | ||
<p align="justify">Since ''Bacillus subtilis'' is not an organism commonly used in iGEM, please check out our [https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction Introduction].</p> | <p align="justify">Since ''Bacillus subtilis'' is not an organism commonly used in iGEM, please check out our [https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction Introduction].</p> | ||
| + | <div class="box"> | ||
==''Bacillus'' Vectors [[File:LMU Backbone.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors|Vectors]]== | ==''Bacillus'' Vectors [[File:LMU Backbone.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors|Vectors]]== | ||
| - | + | {| "width=100%" style="text-align:center;" style="align:right"| | |
| - | + | |<p align="justify">We have generated a suit of BioBrick-compatible vectors: three empty insertional backbones with different antibiotic resistances and integration loci, two reporter and two expression vectors. Here is a list of all the vectors we cloned and used.</p> | |
| - | <p align="justify">We have generated a suit of BioBrick-compatible vectors: three empty insertional backbones with different antibiotic resistances and integration loci, two reporter and two expression vectors. Here is a list of all the vectors we cloned and used.</p> | + | |[[File:LMU-Munich-PSBBs1C.png|200px|right|link=Team:LMU-Munich/Bacillus BioBricks/Vectors]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Bacillus BioBricks/Vectors]] | ||
|} | |} | ||
| - | + | </div> | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 20:41, 24 October 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

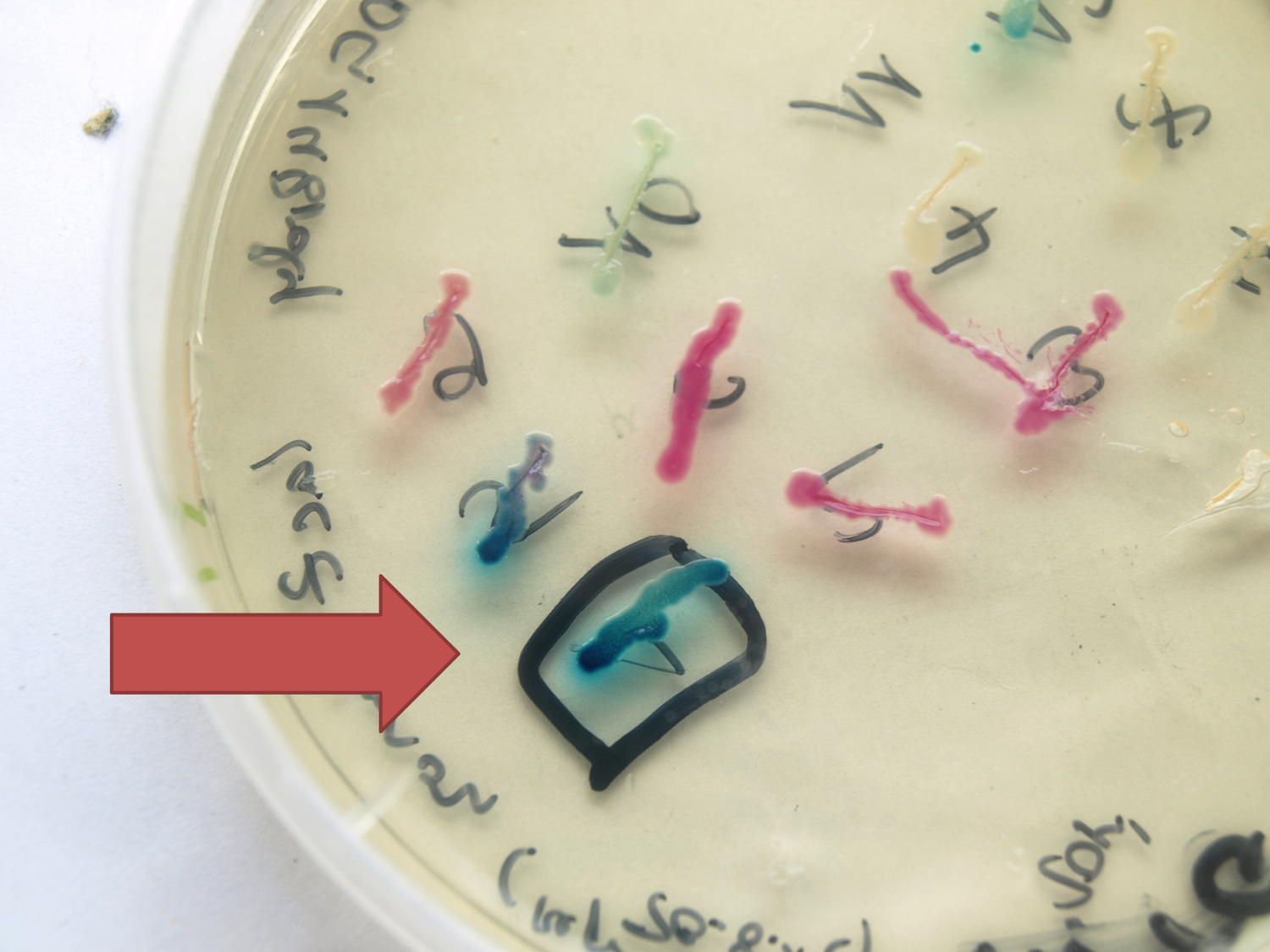
B4 - 22 core parts for Bacillus subtilis
A major goal of our iGEM project is to introduce B. subtilis as a new chassis for BioBrick-based Synthetic Biology. For that purpose, we created a toolbox of Bacillus BioBricks to contribute to the registry to make it accessible to many more future iGEM-Teams! This Bacillus BioBrick Box (B4) contains the following Bacillus specific parts:
| Vectors | Promoters | Reporters | Affinity tags |

| 
| 
| |
|
pSBBs1C |
Anderson |
gfp |
Flag |
Since Bacillus subtilis is not an organism commonly used in iGEM, please check out our Introduction.
Bacillus Vectors 
Bacillus Promoters 
To provide a set of promoters of different strength we characterized several promoters in Bacillus subtilis. They can be divided in three different groups:
- the constitutive promoters from the [http://partsregistry.org/Part:BBa_J23100 Anderson collection] from the Partsregistry
- the constitutive promoters PliaG, Pveg and PlepA from B. subtilis
- the inducible promoters PliaI and xylR-Pxyl from B. subtilis
For the characterization of the different promoters we used the lux operon ![]() . Here, promoter activity leads to expression of the luciferase and hence light production, which can be measured as luminescence.
. Here, promoter activity leads to expression of the luciferase and hence light production, which can be measured as luminescence.
We also used the reporter gene lacZ ![]() . Here, promoter activation results in expression of a β-galactosidase, whose activity can be measured by breakdown of the chromophoric substrate ONPG (β-galactosidase assays). We used the reporter vector pSBBs1C-lacZ. See this page for an overview and background information of all evaluated promoters and see the Data page for more details.
. Here, promoter activation results in expression of a β-galactosidase, whose activity can be measured by breakdown of the chromophoric substrate ONPG (β-galactosidase assays). We used the reporter vector pSBBs1C-lacZ. See this page for an overview and background information of all evaluated promoters and see the Data page for more details.
Overview of all evaluated promoters
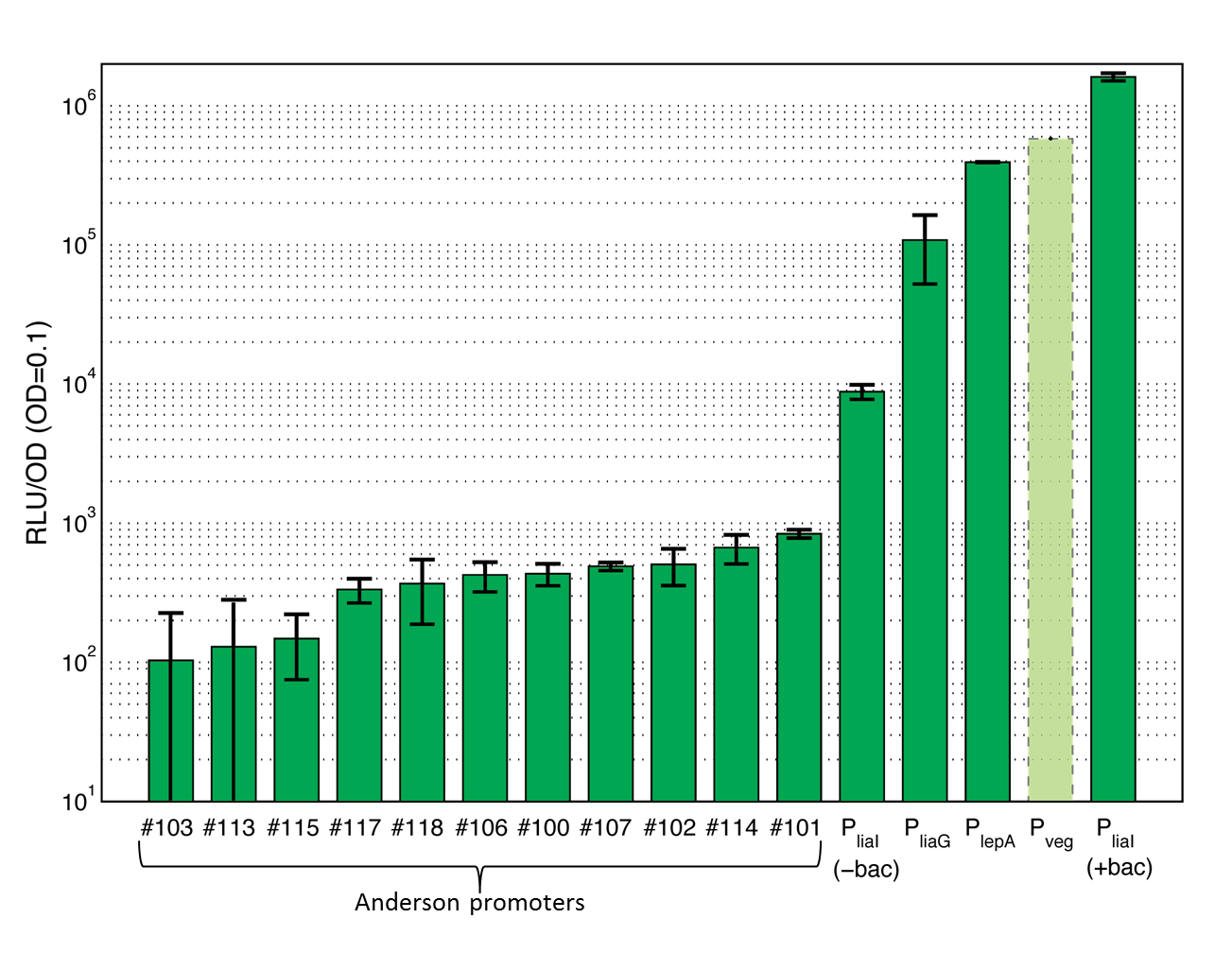
This section gives an overview on the strength of all evaluated promoters, which span a large range of activities. For more details and informations of the experiments see the Data page of the promoters. Note that Pveg was not evaluated with luminescence measurements and this bar is just projected from the results of the β-galactosidase assay.
|
Evaluation of Anderson promoters in B. subtilis
The first group of promoters evaluated are the promoters of the [http://partsregistry.org/Part:BBa_J23100 Anderson collection] ("Anderson promoters"). They have already been extensively measured in Escherichia coli where they all showed a constitutive behavior with different strength. In this project, eleven Anderson promoters were characterized in B. subtilis with the lux operon as a reporter. In B. subtilis these promoters show quiet low activity (see Data Anderson promoters ![]() ).
To confirm these results some Anderson promoters were also evaluated with the reporter gene lacZ by doing β-galactosidase assays (see Data Anderson promoters
).
To confirm these results some Anderson promoters were also evaluated with the reporter gene lacZ by doing β-galactosidase assays (see Data Anderson promoters ![]() ).
).
- J23100 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823004 BioBrick:BBa_K823004])
- J23101 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823005 BioBrick:BBa_K823005])
- J23102 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823006 BioBrick:BBa_K823006])
- J23103 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823007 BioBrick:BBa_K823007])
- J23106 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823008 BioBrick:BBa_K823008])
- J23107 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823009 BioBrick:BBa_K823009])
- J23113 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823010 BioBrick:BBa_K823010])
- J23114 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823011 BioBrick:BBa_K823011])
- J23115 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823012 BioBrick:BBa_K823012])
- J23117 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823013 BioBrick:BBa_K823013])
- J23118 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823014 BioBrick:BBa_K823014])
Constitutive promoters from B. subtilis as novel BioBricks
The second group of promoters are a set of constitutive promoters from B. subtilis that we have added to the Registry. We evaluated the promoters [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823003 Pveg] and [http://partsregistry.org/Part:BBa_K823002 PlepA] using the lux operon as well as the lacZ gene as reporters.
- PliaG ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823000 BioBrick:BBa_K823000])
PliaG is a constitutive promoter from B. subtilis. It is responsible for the transcription of the last four genes of the liaIHGFSR locus and hence for the production of the components of the LiaRS system, which is important for the detection of cell wall antibiotics [http://www.ncbi.nlm.nih.gov/pubmed?term=Journal%20of%20Bacteriology%2C%20188%20%2814%29%3A%205153%E2%80%935166: (Jordan et al., 2006)]. PliaG was evaluated with the lux operon (see Data constitutive promoters ![]() ) as well as the lacZ (see Data constitutive promoters
) as well as the lacZ (see Data constitutive promoters ![]() ) as reporter. This promoter showed a much higher activity than the Anderson promoters, but was still relatively weak in comparison to other evaluated Bacillus promoters.
) as reporter. This promoter showed a much higher activity than the Anderson promoters, but was still relatively weak in comparison to other evaluated Bacillus promoters.
- Pveg ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823003 BioBrick:BBa_K823003])
Pveg is known to show a strong constitutive activity during the vegetative growth phase and sporulation. This promoter is important for the transcription of the veg gene, which plays a role during sporulation [http://www.ncbi.nlm.nih.gov/pubmed?term=J.%20Biochem.%2C%20133%20%284%29%3A%20475%E2%80%93483: (Fukushima et al., 2003)]. Pveg was only measured by using the reporter gene lacZ (see Data constitutive promoters ![]() ). This promoter was the strongest of our evaluation. It failed to clone into the lux reporter vector, presumably because of its strength.
). This promoter was the strongest of our evaluation. It failed to clone into the lux reporter vector, presumably because of its strength.
- PlepA ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823002 BioBrick:BBa_K823002])
PlepA is a constitutive promoter that is important for the transcription of the bicistronic operon. One of the expressed proteins is the protein PlepA [http://www.ncbi.nlm.nih.gov/pubmed?term=Microbiology%2C%20142%3A%201641%E2%80%931649: (Homuth et al., 1996)]. PlepA plays an important role during translation as it can move the mRNA-tRNA complex one step back in the ribosome which is expected to improve the fidelity of translation [http://www.ncbi.nlm.nih.gov/pubmed?term=Cell%2C%20127%20%284%29%3A%20721%E2%80%93733: (Qin et al., 2006)]. This protein was evaluated with the lux operon as a reporter (see Data constitutive promoters ![]() ). The activity of this promoter is between the activity of the strongest Bacillus promoter Pveg and the weak PliaG.
). The activity of this promoter is between the activity of the strongest Bacillus promoter Pveg and the weak PliaG.
Inducible promoters from B. subtilis as a novel BioBrick
The last group of promoters consists of two inducible promoters from B. subtilis, PliaI and xylR-Pxyl. They are useful if the strength and timing of gene expression needs to be tightly controlled, because these promoters need an inducer to initiate transcription. PliaI is evaluated with the reporters lux and lacZ and added this novel promoter to the registry.
- PliaI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823001 BioBrick:BBa_K823001])
PliaI is an inducible promoter from B. subtilis, which responds to antibiotics that interfere with the integrity and biosynthesis of the cell wall [http://www.ncbi.nlm.nih.gov/pubmed/15273097 (Mascher et al., 2004)]. In the presence of a stimulus, the two-component system LiaRS is activated. The activated response regulator LiaR binds to the operator of the promoter and induces the transcription of the lia locus. When the promoter is turned on the two proteins LiaI and LiaH are expressed which play an important role in the stress response of B. subtilis. The major strength of this promoter is its very low basal activity in the absence of an inducer and its high dynamic range [http://www.ncbi.nlm.nih.gov/pubmed/15273097 (Mascher et al., 2004)]. This promoter is evaluated with the reporter lux (see Data inducible promoters ![]() ) as well as lacZ (see Data inducible promoters
) as well as lacZ (see Data inducible promoters ![]() ). The induction was measured with different concentrations of bacitracin, demonstrating a concentration-dependent response over a large range of promoter activities.
). The induction was measured with different concentrations of bacitracin, demonstrating a concentration-dependent response over a large range of promoter activities.
- xylR-Pxyl ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K8230015 BioBrick:BBa_K823015])
Pxyl-xylR is a xylose-inducible promoter from B. subtilis. XylR is the repressor of Pxyl in the absence of the sugar xylose. In the presence of xylose, XylR dissociates from the operator and Pxyl is active (see e.g. [http://www.ncbi.nlm.nih.gov/pubmed/2544559 Kreuzer et al.]). For this promoter we have not yet suceeded to clone it in a reporter vector to evaluate the activity. So far, there is no data for this promoter.
Bacillus Reporters 
We designed and codon-optimized a set of reporters that are commonly used in B. subtilis. All reporters have a modified iGEM Freiburg standard ([http://partsregistry.org/Help:Assembly_standard_25 RFC 25]) pre- and suffix for assembly of in-frame fusion proteins. Our prefix also includes the B. subitlis optimized ribosome binding site.
Find out more about the design of our prefix with ribosome binding site.
prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
suffix: ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
- GFP ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823039 BioBrick:BBa_K823039])
|
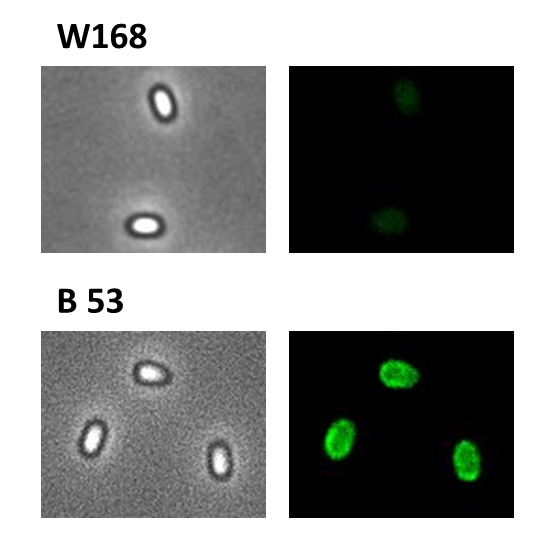
We took a gfp derivate of the Bacillus subtilis plasmid pGFPamy and added the BioBrick compatible pre- and suffix of the Freiburg standard ([http://partsregistry.org/Help:Assembly_standard_25 RFC 25]). The functionality of this BioBrick verified by creat e.g. our Sporobeads.
- mKate2 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823029 BioBrick:BBa_K823029])
|
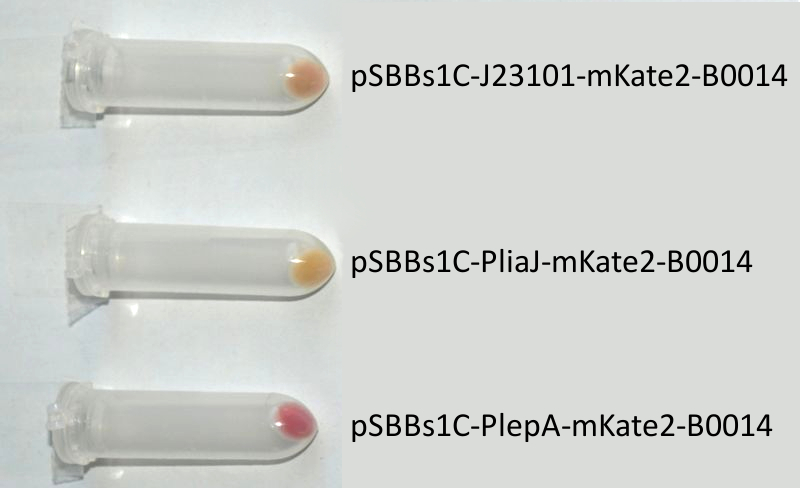
We codon-optimized the sequence of this monomeric far-red fluorescence protein for the use in B. subtilis and synthesized it (by [http://de-de.invitrogen.com/site/de/de/home/Products-and-Services/Applications/Cloning/gene-synthesis.html GeneArt]) with pre- and suffix of the Freiburg standard. We cloned this reporter in front of the terminator B0014. For the evaluation, this reporter was successfully combined with the promoters PliaI, PlepA and the Anderson promoter J23101 in the empty Bacillus vector pSBBs1C from our BacillusBioBrickBox. At the moment we already have the right construct integrated into the chromosome of B. subtilis. Unfortunately, we have not yet measured this reporter.
Reference: [http://www.evrogen.com/products/mKate2/mKate2.shtml mkate2]
- LacZ ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823019 BioBrick:BBa_K823019])
|
This evaluated lacZ gene is derived from the Bacillus reporter vector pAC6. It is constructed in the Freiburg Standard ([http://partsregistry.org/Help:Assembly_standard_25 RFC 25]) for in-frame fusion proteins. It also includes a ribosome binding site optimized for Bacillus subtilis translation. This lacZ BioBrick was tested in the expression vector pSBBs0K-Pspac. This construct gave output, so the reporter gene is functional qualitatively. See Data in the vector evaluation section of pSBBs0K-Pspac. A more qualitatively measurement will follow soon.
Reference for the vector: [http://www.ncbi.nlm.nih.gov/pubmed/11902727 pAC6]
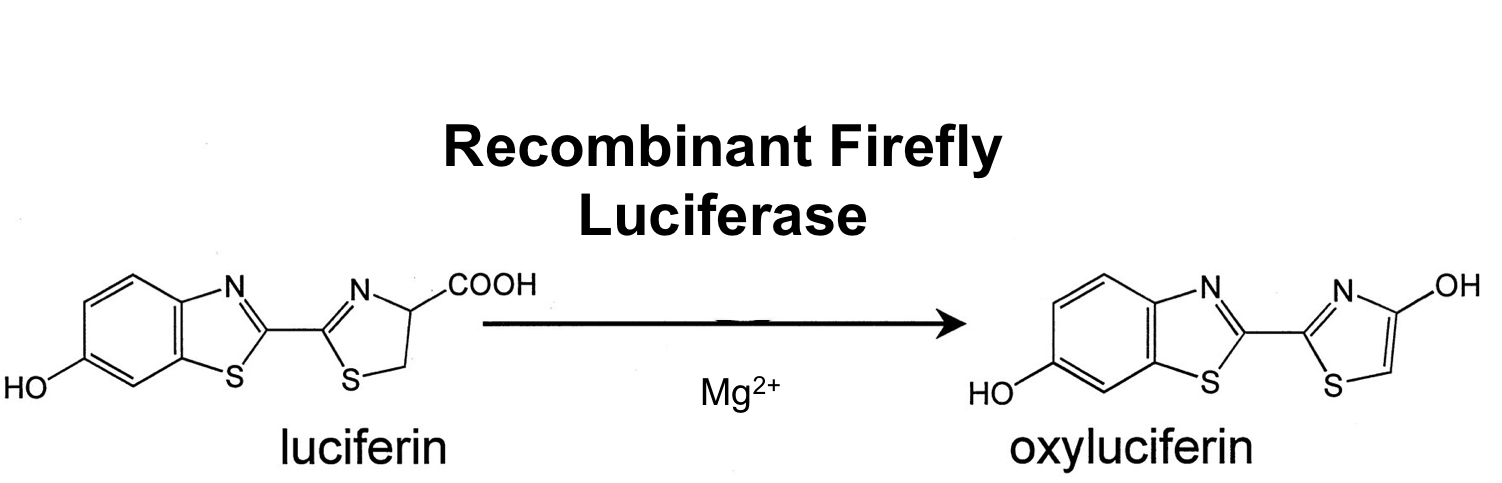
- luc ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823028 BioBrick:BBa_K823028])
The luc gene encodes the monomeric firefly luciferase, which requires a special substrate to produce luminescence. We codon-optimized it for Bacillus subtilis, added a ribosome binding site and synthesized by [http://de-de.invitrogen.com/site/de/de/home/Products-and-Services/Applications/Cloning/gene-synthesis.html GeneArt].
Firefly luciferase is by far the most commonly used bioluminescent reporter. This monomeric enzyme of 61kDa catalyzes a two-step oxidation reaction to yield light, usually in the green to yellow region, typically 550–570nm . The first step is activation of the luciferyl carboxylate by ATP to yield a reactive mixed anhydride. In the second step, this activated intermediate reacts with oxygen to create a transient dioxetane that breaks down to the oxidized products, oxyluciferin and CO2. Upon mixing with substrates, firefly luciferase produces an initial burst of light that decays over about 15 seconds to a low level of sustained luminescence. This kinetic profile reflects the slow release of the enzymatic product, thus limiting catalytic turnover after the initial reaction.
The popularity of native firefly luciferase as a genetic reporter is due to the sensitivity and convenience of the enzyme assay and tight coupling of protein synthesis with enzyme activity. Firefly luciferase, which is encoded by the luc gene, is a monomer that does not require any post-translational modifications; it is available as a mature enzyme directly upon translation of its mRNA. Catalytic competence is attained immediately after release from the ribosome. Also, luciferase has a very short half-life in cells (approximately 3 hours). Combined, these properties make luciferase an extremely responsive reporter, far more so than other commonly used reporters.
|
It was used in B. subtilis before ([http://www.ncbi.nlm.nih.gov/pubmed/21552330 Mirouze et al. 2011]). The non-optimized version of this reporter gene was already successfully used in B. subtilis. While we have not found the time to evaluate this BioBrick we expect that it will work at least as good.
Affinity Tags 
All our tags have been synthesized by [http://de-de.invitrogen.com/site/de/de/home/Products-and-Services/Applications/Cloning/gene-synthesis.html GeneArt]. They are designed in Freiburg standard with an optimized ribosome binding site upstream. We have not yet tested our tags.
Find out more about the design of our prefix with ribosome binding site.
prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
suffix: ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
- 3x Flag-tag [http://partsregistry.org/Part:BBa_K823034 (BioBrick:BBa_K823034)]
The Flag-tag was the first epitope tag to be published ([http://www.nature.com/nbt/journal/v6/n10/full/nbt1088-1204.html Hopp, Prickett et al. (1988)]). It consists of eight hydrophobic amino acids: DYKDDDDK. To enhence senstitivity in Western blots, it is routinely used as a 3x Flag tag in B. subtilis [http://www.ncbi.nlm.nih.gov/pubmed?term=wiegert%20flag (Kaltwasser et al., 2002)]. Its sequence is: DYKDHDGDYKDHDIDYKDDDDK. There are a variety of monoclonal antibodies against this tag, N-terminal as well as position insensitive.
- HA - tag [http://partsregistry.org/Part:BBa_K823035 (BioBrick:BBa_K823035)]
The HA-tag is an epitope derived from the HA-virus. There was first an antibody against it and then the epitope was characterized ([http://www.ncbi.nlm.nih.gov/pubmed/6204768 Wilson, I.A. et al. (1984)]). It was then furthermore used as a tag for protein purification and recognition ([http://www.ncbi.nlm.nih.gov/pubmed/2455217 Field et al. (1988)]). The amino acid sequence is: YPYDVPDYA.
- cMyc - tag [http://partsregistry.org/Part:BBa_K823036 (BioBrick:BBa_K823036)]
The cMyc-tag is derived from the cMyc gene product. Antibodies were generated from the immunization with synthetic peptides from the cMyc sequence ([http://mcb.asm.org/content/5/12/3610.short Evan, Bishop et al.(1985)]). The amino acid sequence is EQKLISEEDL.
- His - tag [http://partsregistry.org/Part:BBa_K823037 (BioBrick:BBa_K823037)]
The His-tag is a metal chelating peptide ([http://www.nature.com/nbt/journal/v6/n11/full/nbt1188-1321.html Hochul,Stüber et al. (1988)]) consisting of at least 6 histidin residues. It can be used for protein purification by metal2+ion-containing columns (nickel). There are also antibodies against this tag, or nickel/cobalt containing fluorescent probes can be used for detection. Moreover, an immobilization is possible in nickel/cobalt coated plastikware. The aminoacid sequence is:HHHHHHHHHH
- Strep - tag [http://partsregistry.org/Part:BBa_K823038 (BioBrick:BBa_K823038)]
The Strep-tag is a mimicry peptide of biotin which binds to Streptavidin ([http://www.sciencedirect.com/science/article/pii/S1050386299000339 Skerra, Schmidt (1999)]). Its sequence is WSHPQFEK. It can be used for protein purification, immobilization with streptavidin or strep-tactin ([http://www.ncbi.nlm.nih.gov/pubmed/9415448 Voss, Skerra (1997)]) or detection with Strep-tactin or antibodies.

| 
| 
| 
|
| Bacillus Intro | Bacillus BioBrickBox | Sporobeads | Germination STOP |
 "
"