Team:SJTU-BioX-Shanghai/Project/project1.3
From 2012.igem.org
AleAlejandro (Talk | contribs) (→RNA Signal) |
AleAlejandro (Talk | contribs) (→RNA Signal) |
||
| Line 55: | Line 55: | ||
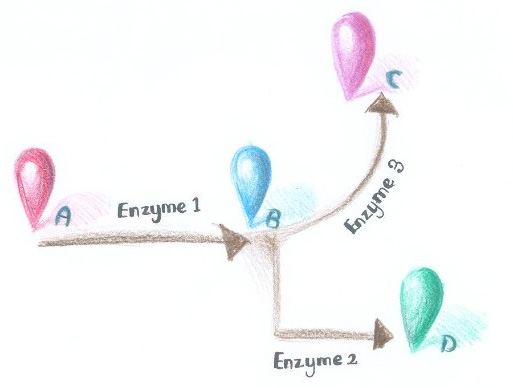
| - | So far, ''Membrane Accelerator'' and ''Membrane Rudder'' are both post-translation controlling device to regulate metabolic flux of the host cell. To connect this relatively isolated post-translation control system to genetic circuits, we employed RNA signal, which is present in cytoplasm. When certain RNA molecule with two aptamer domains is present in cells, their cognate aptamer binding proteins can thus | + | So far, ''Membrane Accelerator'' and ''Membrane Rudder'' are both post-translation controlling device to regulate metabolic flux of the host cell. To connect this relatively isolated post-translation control system to genetic circuits, we employed RNA signal, which is present in cytoplasm. When certain RNA molecule with two aptamer domains is present in cells, their cognate aptamer binding proteins can thus aggregate together. |
[[Image:12SJTU RNASIGNAL.jpg|thumb|400px|center|''Fig.3'' : | [[Image:12SJTU RNASIGNAL.jpg|thumb|400px|center|''Fig.3'' : | ||
<br> | <br> | ||
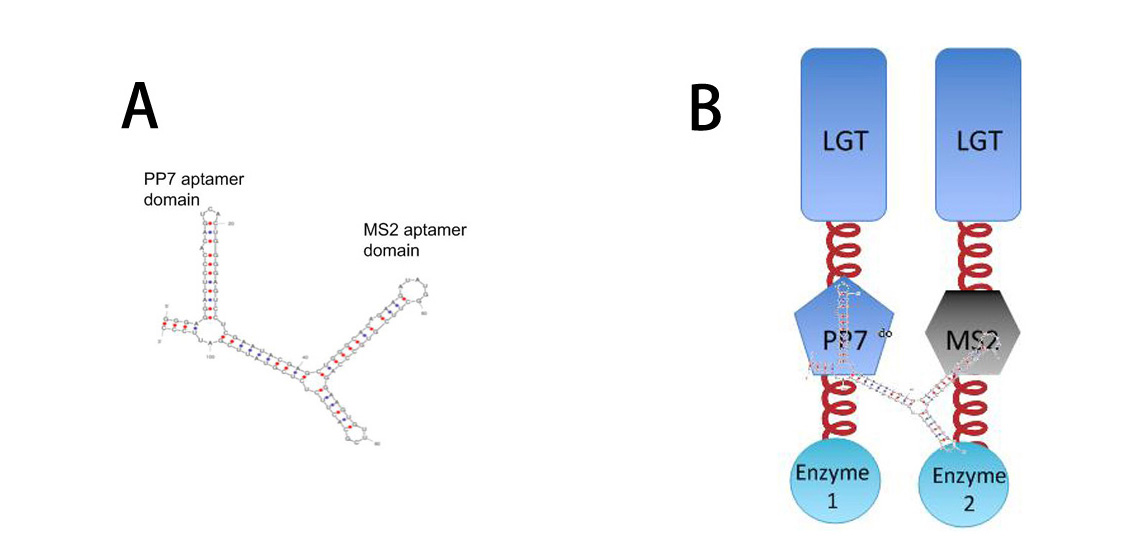
| - | '''A''': Sketch of signal RNA molecule (called D0) which consists of PP7 and MS2 aptamer domains. | + | '''A''': Sketch of signal RNA molecule (called D0) which consists of PP7 and MS2 aptamer domains. PP7 aptamer binding protein could dimerize with PP7 RNA aptamer domain; MS2 aptamer binding protein could dimerize with MS2 RNA aptamer domain. |
<br> | <br> | ||
'''B''': Through fusing PP7 and MS2 aptamer binding protein to Membrane Anchor, RNA molecule in Figure A can function as a bridge to connect different proteins, thus decreasing distance between corresponding proteins.]] | '''B''': Through fusing PP7 and MS2 aptamer binding protein to Membrane Anchor, RNA molecule in Figure A can function as a bridge to connect different proteins, thus decreasing distance between corresponding proteins.]] | ||
| - | + | By now, if we want to assemble certain proteins through external signal, we need to find corresponding signal-induced dimer or oligomer. But if we place RNA D0 (with PP7 and MS2 aptamer domains) under various promoters regulated by different signals, approaches to induce dimerization would be expanded sharply. Thus, ''Membrane Rudder'' could sense much more signals. | |
We have constructed the PP7 and MS2 standardized BioBrick part, [http://partsregistry.org/Part:BBa_K771111 Part:BBa_K771111] and [http://partsregistry.org/Part:BBa_K771112 Part:BBa_K771112], respectively. | We have constructed the PP7 and MS2 standardized BioBrick part, [http://partsregistry.org/Part:BBa_K771111 Part:BBa_K771111] and [http://partsregistry.org/Part:BBa_K771112 Part:BBa_K771112], respectively. | ||
Revision as of 14:10, 24 October 2012
| ||
|
 "
"