Team:SJTU-BioX-Shanghai/Project
From 2012.igem.org
AleAlejandro (Talk | contribs) (→Why MEMBRANE?) |
AleAlejandro (Talk | contribs) (→Introduction) |
||
| Line 46: | Line 46: | ||
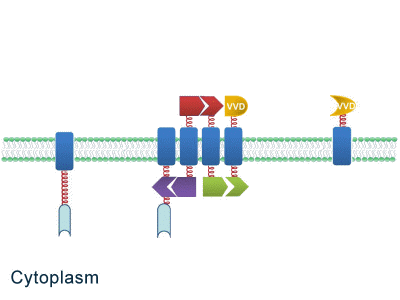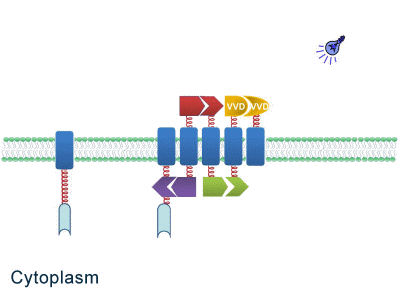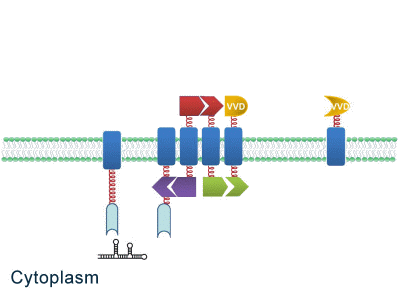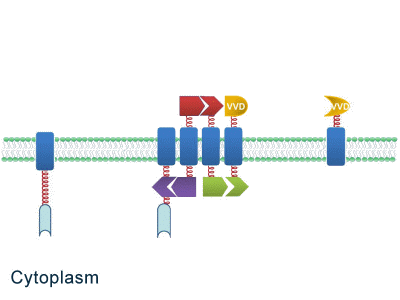
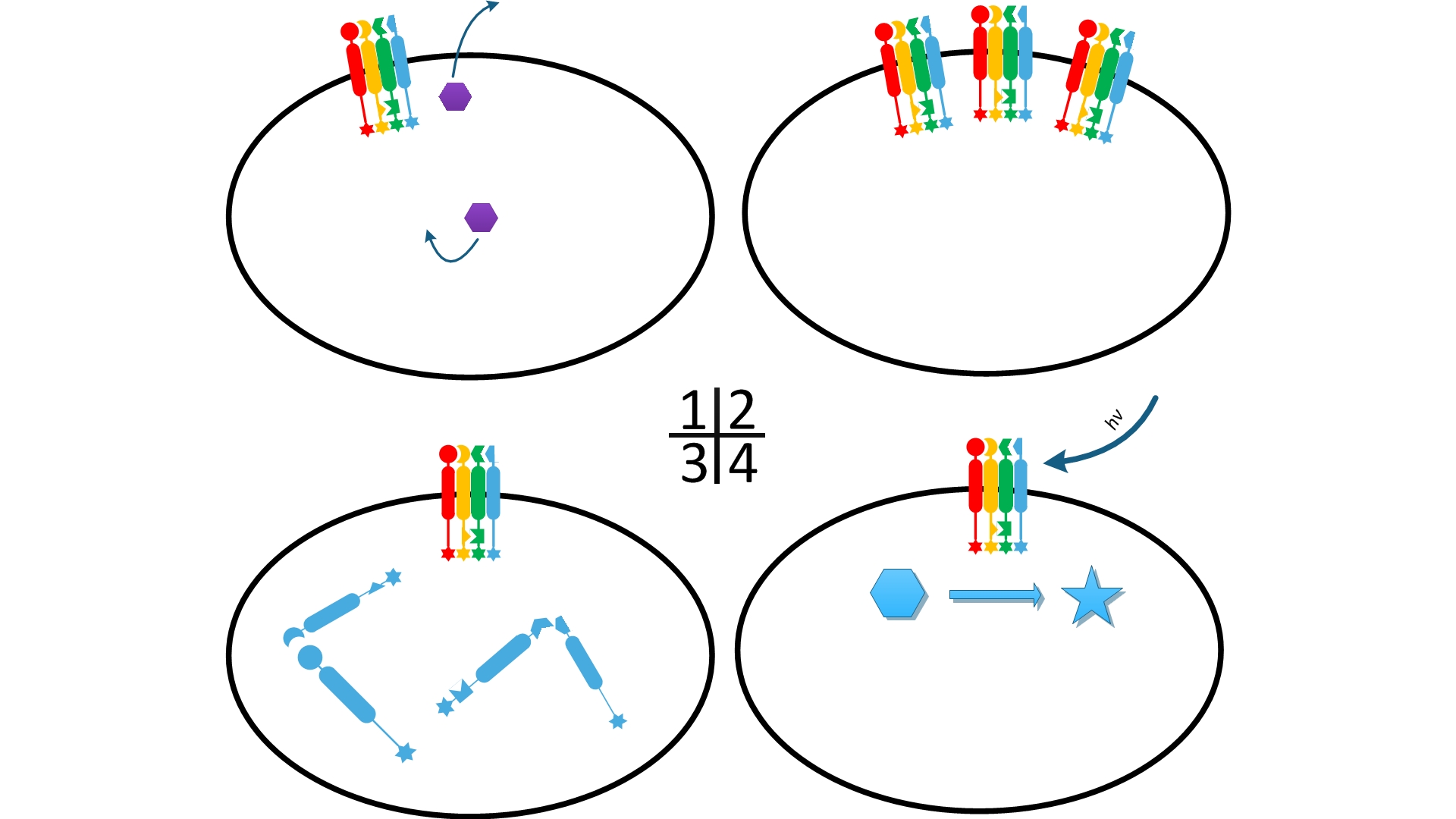
One of our devices, called ''Membrane Accelerator'', functions by localizing and organizing enzymes on membrane surface. ''E.coli'' inner membrane serves as a two-dimensional plane that can accommodate various protein assemblies linked with enzymes. Otherwise diffusing enzymes can form clusters on membrane through interacting protein domains and ligands. Enzyme clusters help substrates flow between enzymes, and thus increase yields of sequential biological reactions. We not only applied the ''Membrane Accelerator'' into biosynthetic pathway but also biodegradation pathway, which is proposed for the first time in synthetic biology. Previous researches on scaffold system all focused on biosynthesis. | One of our devices, called ''Membrane Accelerator'', functions by localizing and organizing enzymes on membrane surface. ''E.coli'' inner membrane serves as a two-dimensional plane that can accommodate various protein assemblies linked with enzymes. Otherwise diffusing enzymes can form clusters on membrane through interacting protein domains and ligands. Enzyme clusters help substrates flow between enzymes, and thus increase yields of sequential biological reactions. We not only applied the ''Membrane Accelerator'' into biosynthetic pathway but also biodegradation pathway, which is proposed for the first time in synthetic biology. Previous researches on scaffold system all focused on biosynthesis. | ||
| - | |||
| - | |||
| - | |||
| - | ''Fig.1:'' Sketch of ''Membrane Accelerator'' | + | [File:12SJTU membrane accelerator sketch.jpg|400px|center|''Fig.1:'' Sketch of ''Membrane Accelerator''} |
Although some work has been done in reaction acceleration, it has always been a challenge to artificially and dynamically control those reactions. Our ''Membrane Rudder'' device, however, offers a novel method to control the direction of biochemical reactions through varieties of signals. We further combined the whole post-translational control system with genetic circuits by recruiting RNA aptamer and its corresponding binding protein. Thus RNA signal could also be recruited to dynamically control biochemical reaction. | Although some work has been done in reaction acceleration, it has always been a challenge to artificially and dynamically control those reactions. Our ''Membrane Rudder'' device, however, offers a novel method to control the direction of biochemical reactions through varieties of signals. We further combined the whole post-translational control system with genetic circuits by recruiting RNA aptamer and its corresponding binding protein. Thus RNA signal could also be recruited to dynamically control biochemical reaction. | ||
Revision as of 06:03, 24 October 2012
| ||
|
 "
"