Team:UC-Merced/Notebook
From 2012.igem.org
Darealsunny (Talk | contribs) |
Darealsunny (Talk | contribs) |
||
| Line 46: | Line 46: | ||
<tr> | <tr> | ||
| - | <td align=center><font color=black>September 26, 2012:<br> | + | <td align=center><font color=black><h4>September 26, 2012:</h4><br> |
Glycerol stocks + Agar+ Nanodrop</font></td> | Glycerol stocks + Agar+ Nanodrop</font></td> | ||
<td align=left><font color=black> | <td align=left><font color=black> | ||
Revision as of 02:32, 4 October 2012
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Notebook | Safety | Background | Attributions |
|---|
September 19, 2012:DNA Extraction |
Data: 215 Cells/5 Squares = 43 cells/square x = 43*100*4*10^(-3) x = 43 cells/square (43/.004 mm^3) 1000100 = 1.075*10^9 cells/mL (2.5*10^(7)) cells / (1.075*10^9 cells) = .023mL of bacteria Then start the extraction using the mini-prep kit (insert link to mini prep kit protocol ) Turn on the thermomixer when the buffers are being added -Temp at 56 degree C for 10 minutes -Takes around 5 minutes to heat up Use filter tips since we’re working with DNA As a funny mess up, we ended up throwing away the glass slip for the hemocytometer!!! |
September 26, 2012:Glycerol stocks + Agar+ Nanodrop |
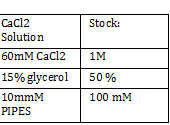
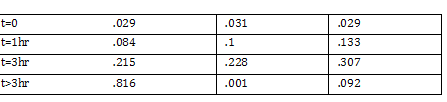
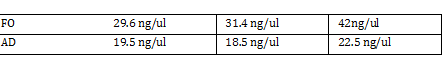
Nanodrop -add small drop of DiH2O 3 times -dab with kim wipe do not wipe -dab both top and bottom bend -log onto ND1000 program and select nucleic acid -add DIH2O with micropipette and close top 2x - add 1 micro liter of DNA to nanodrop, close and measure -save and print Results: 24.1 nanogram/microliter 260/280 = 1.82 260/230 = .49 Remember to clean machine with DI H2O after use! 1 M CaCl2 Protocol -calculate morality of 1M CaCl2 Mass = M*Vol*MW Mass = 14.702 grams per 100 mL of H2O Add 60 L H2O to beaker Add 14.702g CaCl2 to beaker and stir Add back to column and fill up to 100mL Use 150 mL filter until PIPES protocol Weight out 2.307 g of pipes solid Required solution must be at 7pH so add NaOH and measure pH using pH stripes or indicator Once at 7 pH add dH2O until 100mL Use 150 mL filter unit |
September 27, 2012:Antibiotic plates |
Calculations (Note: Dilution formula was used) Chloramphenicol: (6mg/mL)x=(.025mg/mL)(250mL) x = 1mL Kanamycin: (25 mg/mL)x = (0.05 mg/mL)(250mL) x = .5mL Streptomycin: (20mg/mL)x = (0.02 mg/mL)(250mL) x = .25 mL Antibiotic Plates (Con’t): Data 1. Chloramphencial: Recommended = 25microgram/mL, stock = 6 mg/mL 2. Kanamycin: Recommended = 50 microgram/mL, stock = 25 mg/mL 3. Strephtomycin: Recommended = 20 microgram/mL, stock = 20 mg/mL Note: Color coded the plates: Chloramphenciol (Green) Kanamycin (Orange) Strep/Chloram (Green Black) Bacterial Incubation with Antibiotics E coli W – no antibiotics FMJ 39- strepromycin (25microgram/mL) JW 228- Kanamycin (25 microgram/mL) Bba_K27300- 1000x Amp To make 1000x Amp (25mg/mL)V = (25microgram/mL)(5mL) V = 5 microliter Incubate bacteria in 5mL of LB with antibiotics Glycerol stocks of bacteria Note: Remember to use Aseptic Technique! -Mix 700 micro liter log phase culture with 300 microliter 50 % glycerol -Vortex -Store into cryotube -place in -80 degree C for storage Nanodrop for HTCC 2181 -Same procedure as the one above Results: -260/280: 1.78 -260/230: .17 -2 ng/microliter 260/280: 1.98 260/230: .13 1.4 ng/microliter 260/280: 2.53 260/230: .13 2.9 ng/microliter TAE buffer 1 x for electrophoresis Calculations C1V1 = C2V2 50V1 = (1x)(1000mL) V1 = 20mL of 50x Primer Stocks [100 micromolar primered stocks] -AD BB Pfx Add 633 microliter DH20 -AD GSA Bt Add 1213 micro lite rDH2O -FO GSA Tp Add 1314 microliter DH2O FO BB Sfx Add 615 microliter DH2O Diluted to 10 micromolar working volumes Incubated FMJ39 and JW 1228 for competent because not sure which strain will be used |
 "
"