Team:British Columbia/Pathway
From 2012.igem.org
(Difference between revisions)
| Line 14: | Line 14: | ||
<p align=center><img src="https://static.igem.org/mediawiki/2012/9/9c/UbcigemSlide1.jpg"></p> | <p align=center><img src="https://static.igem.org/mediawiki/2012/9/9c/UbcigemSlide1.jpg"></p> | ||
| - | < | + | <h2>Modeling Metabolism:</h2> <i>Rhodococcus erythropolis</i> and <i>Pseudomnas fluorescens</i> are often both prevalent in similar niches. With a high likelihood that these organisms encounter each other, studies have been conducted to assess their metabolic properties in co-culture. In a study by Kayser et al, it was shown that the 4-S biodesulferization pathway in <i>R. erythropolis</i> was more active in the presence of <i>P. fluorescens</i> [2]. As <i>P. fluorescens</i> does not biodesulferize DBT and sulfate is known to repress the 4-S pathway, we looked to analyze the genomes of the two organisms in the context of sulfur metabolism. </br></br> |
The 4-S pathway releases sulfite, which is toxic to the cell, therefore genomes were analyzed with the aforementioned pipeline for pathways involved in metabolizing sulfite. It was found that both organisms have annotated genes converting sulfite into sulfate via a reductase; however, the organisms differ on the downstream metabolism of the sulfate. Where both organisms have an assimilatory sulfate metabolism, only <i>P. fluorescens</i> has a dissimilatory sulfate metabolism (Figure 2, 3). Based on these findings, we can hypothesize that <i>R. erythropolis</i> and <i>P. fluorescens</i> likely excrete and catabolize any excess sulfate, respectively. This provides an explanation for the improved desulfurization found in co-culture conditions. <i>P. fluorescens</i> potentially removes sulfate from the environment allowing increased secretion by <i>R. erythropolis</i> and thereby removing suppression of the 4-S pathway. The prediction of distributed sulfite metabolism to improve biodesulferization in the environment provides both a testable hypothesis and direction in improving biodesulferization through synthetic pathway distribution. </br></br> | The 4-S pathway releases sulfite, which is toxic to the cell, therefore genomes were analyzed with the aforementioned pipeline for pathways involved in metabolizing sulfite. It was found that both organisms have annotated genes converting sulfite into sulfate via a reductase; however, the organisms differ on the downstream metabolism of the sulfate. Where both organisms have an assimilatory sulfate metabolism, only <i>P. fluorescens</i> has a dissimilatory sulfate metabolism (Figure 2, 3). Based on these findings, we can hypothesize that <i>R. erythropolis</i> and <i>P. fluorescens</i> likely excrete and catabolize any excess sulfate, respectively. This provides an explanation for the improved desulfurization found in co-culture conditions. <i>P. fluorescens</i> potentially removes sulfate from the environment allowing increased secretion by <i>R. erythropolis</i> and thereby removing suppression of the 4-S pathway. The prediction of distributed sulfite metabolism to improve biodesulferization in the environment provides both a testable hypothesis and direction in improving biodesulferization through synthetic pathway distribution. </br></br> | ||
Revision as of 02:24, 4 October 2012

Our Pathway Model
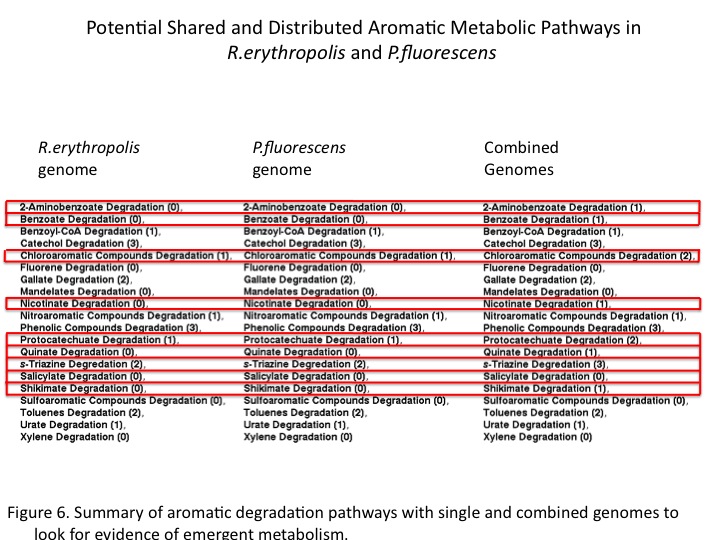
The study of environmental genomics attempts to capture the taxonomic and functional diversity of natural microbial communities. Our host at UBC, the Hallam lab, designs novel tools for analyzing the gene content in the context of distributed metabolism. Recently, a pipeline has been developed for the automated construction and visualizing of metabolic pathways from genomic data by integrating software such as Pathway Tools, Pathologic and Metacyc [1]. This provided us an opportunity to model pathway compartmentalization and distribution amongst microbes in the natural environment as it applies to our project.Summary of the pipeline for community-level metabolic analysis (Figure 1):
First, open reading frames predicted from sequence data using Prodigal are annotated by protein BLAST and summarized in a GenBank file. Pathway/genome databases (PGDBs) are generated from sequence data in a manner that does not constrain predictions within the scope of model organisms. This allows for a community-based analysis that can be visualized using Pathway Tools.
Modeling Metabolism:
Rhodococcus erythropolis and Pseudomnas fluorescens are often both prevalent in similar niches. With a high likelihood that these organisms encounter each other, studies have been conducted to assess their metabolic properties in co-culture. In a study by Kayser et al, it was shown that the 4-S biodesulferization pathway in R. erythropolis was more active in the presence of P. fluorescens [2]. As P. fluorescens does not biodesulferize DBT and sulfate is known to repress the 4-S pathway, we looked to analyze the genomes of the two organisms in the context of sulfur metabolism. The 4-S pathway releases sulfite, which is toxic to the cell, therefore genomes were analyzed with the aforementioned pipeline for pathways involved in metabolizing sulfite. It was found that both organisms have annotated genes converting sulfite into sulfate via a reductase; however, the organisms differ on the downstream metabolism of the sulfate. Where both organisms have an assimilatory sulfate metabolism, only P. fluorescens has a dissimilatory sulfate metabolism (Figure 2, 3). Based on these findings, we can hypothesize that R. erythropolis and P. fluorescens likely excrete and catabolize any excess sulfate, respectively. This provides an explanation for the improved desulfurization found in co-culture conditions. P. fluorescens potentially removes sulfate from the environment allowing increased secretion by R. erythropolis and thereby removing suppression of the 4-S pathway. The prediction of distributed sulfite metabolism to improve biodesulferization in the environment provides both a testable hypothesis and direction in improving biodesulferization through synthetic pathway distribution.



 "
"