Team:Cornell/Notebook/Wetlab
From 2012.igem.org
(→September 3rd) |
(→September 7th) |
||
| Line 636: | Line 636: | ||
Overnight cultures from yesterday looked slightly opaque, but not opaque enough to miniprep from, so Caleb subcultured the cultures hoping the cells would divide fast enough to be ready for miniprepping tomorrow. | Overnight cultures from yesterday looked slightly opaque, but not opaque enough to miniprep from, so Caleb subcultured the cultures hoping the cells would divide fast enough to be ready for miniprepping tomorrow. | ||
| + | |||
| + | Synthetic River Media: | ||
| + | |||
| + | Mark started overnight cultures of all engineered strains to test, again, how growth in synthetic Athabasca river water supplemented with sodium lactate. | ||
===September 8th=== | ===September 8th=== | ||
Revision as of 03:16, 28 September 2012
Contents
|
Overview
Overview We began the summer by holding a synthetic biology bootcamp at the DeLisa Lab. The purpose of this bootcamp was both to introduce new members to techniques in molecular biology and to get a running start on the cloning work for our project. During bootcamp, we successfully constructed both versions of our arsenic reporter, and attempted a Gibson assembly of a naphthalene-degrading plasmid—without success.
In late June, we transitioned from bootcamp to our permanent bench space in Dr. Archer’s lab in Weill Hall. After spending a few weeks setting up the lab space troubleshooting general issues, we successfully constructed both versions of our salicylate reporter and began an alternative approach to construct a plasmid with a naphthalene-degrading operon. In parallel, we realized that electroporation efficiency for Shewanella transformation is less than optimal—to say the least. However, we were able to conjugate our constructs into Shewanella using a protocol provided by Dr. Gralnick.
…
[Transition to work during school year.]
June
June 12th-16th
June 12th, Tuesday
Click to view details.
June 13th, Wednesday
- Set up PCR for four fragments of the nah operon as well as the pBMT-1 backbone, using primers designed to mutate cut-sites concurrently with Gibson Assembly. Also set up a PCR for the entire nah operon. Due to length of the fragments, a longer extension time was chosen.
- If Gibson Assembly fails, but we are able to PCR the entire 10kb nah operon, an alternate method of mutation using three sequential site-directed mutageneses will be pursued.
- Work done by: Caleb, Claire, Dylan, Spencer, Steven, and Swati
June 14th, Thursday
- PCR only amplified nah1, nah3, and nah4 - longer fragments (nah2, the full nah operon, and pBMT-1) were not identified on the PCR product gel. Set up another PCR with shorter extension time.
June 15th, Friday
- PCR of four out of five products visible on gel. Set up PCR of pBMT-1, final fragment required for Gibson Assembly of the nah operon.
June 16th, Saturday
June 17th-23rd
Summary for this week: We tentatively completed Gibson assembly of the nah operon into the pBMT-1 backbone (minus the BioBrick notI and pstI cut sites)and transformed it into E. coli. The undigested Gibson-assembled plasmid ran at an expected size on a gel (the extra bands were rationalized to be various forms of the plasmid due to nicks and supercoiling).
June 17th, Sunday
Today we did PCR of pBMT-1 to make the backbone Gibson assembly compatible.
June 18th, Monday
June 19th, Tuesday
Dylan an Swati ran Gibson Assembly of nah operon fragments to assemble them into the pBMT-1 backbone and then transformed the Gibson assembly product into DH5a electrocompotent E. coli cells.
June 20th, Wednesday
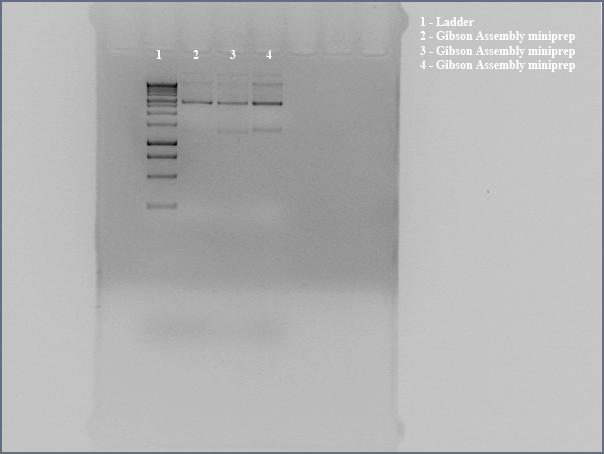
Dylan and Swati set up a digestion of the PCR of Gibson Assembly products and ran the digested products on a gel to see if Gibson worked. Three colonies were observed on the plates of DH5a transformed with Gibson Assembly product, so we made liquid cultures to miniprep and sequence.
June 22nd, Friday
We miniprepped directly transformed Gibson Assembly product to send in for sequencing using the the Gibson nah1F and nah4R primers (each w/ 20 bp overhangs). Before sending in for sequencing, we ran undigested miniprep with gel electrophoresis, looking for large bands corresponding to supercoiled DNA. The gel showed distinct bands for all three lanes, so we interpreted this to mean that we got product. Submitted for sequencing
June 23rd, Saturday
June 24th-30th
June 24th, Sunday
June 25th, Monday
June 26th, Tuesday
June 27th, Wednesday
- Ran digest of gibson-assembled nah operon with gel electrophoresis
June 28th, Thursday
- Gel purified arsR construct
June 29th, Friday
- Vent PCR at 11:00 (DPW)
- Amplifying both previous Phusion PCR band and original p21 template
- Dylan's magic triple anneal method (55/60/63)
- Gel purified PCR product from Phusion template (~1:20 pm) (DPW)
- Quantified product at 22.4 ng/uL
- Set up digestion of p21 PCR product with EcoRI-HF and AscI (~9:00 pm).
- 22 ng/uL --> 45.5 uL sample for 1 ug digest
- Buffer 4
- Ran digestion on gel. (~11:00 pm)
- Sliced out relevant band on gel, stored overnight at -20.
- Miniprepped overnight cultures of PL14-PL20 (~1:00 pm, STC)
- Using C1015 rotor, 6666 x g, the Corning culture tubes only fit in the centrifuge with the lids off
- Made LB, 3x 60 mL in 100 mL bottles (~ 3:00pm, SS)
- Made LB Agar, 4 x 250 mL LB Agar in 500 mL bottles (~3:00, CS)
- Autoclaved LB, LB Agar, and milliQ (~3:30 pm, SS)
- Made LB plates with Kan (~6:30 pm, CMR)
- CUGEM movie outing at 8:00 pm.
- Phusion PCR at 10:00 pm (DPW)
- Dylan's magic triple anneal method (57/65/70, 35 cycles total)
- Amplifying nah operon from Gibson 1
- Appending BioBrick cutsites for ligation into pSB3C5
June 30th, Saturday
- Took PCR out of thermal cycler at 9:00 am (DPW)
- Set up gel using NEB 100bp and 2-log ladders (10:00am)
- Gel extracted PCR product, quantified at ~10ng/uL
- Set up new Phusion PCR using Gibson 1 as template
- Dylan's magic three-anneal method (57.6/65/72)
- Extension time of 3 min.
- Continued gel extraction of p21 PCR digest from previous day (SS)
- Set up ligation of p21 PCR digest extraction and dephosphorylated pBBRBB+mtrB (11:11am, SS)
- Desalted ligation using Millipore membrane paper
- Transformed 2 electrocompetent DH5alpha stocks at 5:30 pm and 5:50 pm, respectively.
- Observed time constants for electroporation of 4.38 ms and 4.24 ms, respectively
- Let cells recover for 1 hour, plated on LB + Kan.
- Set up two ligations of pSB3C5 into PNNL electrocompetent Shewanella strain JG700 (6:30 pm, Sp.C and St.C)
- First transformation performed at usual PNNL voltage of 0.75 kV (time constant of 9.30 ms)
- Second transformation performed at Myers and Myers specification of 0.55 kV (time constant of 9.34 ms).
July
July 1st-7th
A lot of troubleshooting occurred this week. Once we figured out that SYBR Green was causing our gels to run strangely, we switched to staining with EtBr after running the gel. This approach gave us better results. We also continued to try electroporation protocols for transforming Shewanella, and continued to work on PCRing out the salicylate-sensing region.
July 1st, Sunday
Dylan and Caleb set up two gels for electrophoresis. Caleb's was 1% agarose in BIO-RAD Mini-Sub Cell system for continuation of ladder test using SYBR Green, containing NEB 100 bp ladder, NEB 2-log ladder, Promega 1 kb ladder and run at 100V. Dylan's was 1% agarose in Owl box using ethidium bromide, containing the nah operon PCR product from previous night, and run at 55 V.
Our plates of DH5a transformed with p21 ligated into pBBRBB with mtrB grew only one colony, possibly contamination. Dylan ran a colony PCR and got a smear, suggesting that the PCR of p21 or the ligation did not work.
Because we learned that our SYBR Green was causing ladder to run strangely, Dylan decided to redo a Vent PCR to amplify the salicylate reporter region out of p21.
Also, a liquid culture of JG700 was prepared, as well as replating of p21, p22, JG700, JG1220, JG1537, JG1219. (See: strain list)
July 2nd, Monday
Today, Dylan and Caleb ran a gel of a Vent PCR of p21 (the PCR product being the salicylate reporter) at 55 V. Additionally, Caleb decided to run a control gel at 100 V to determine whether higher voltage was a factor in our previous ladder problems (in addition to the SYBR Green stock). While we determined that the higher voltage did not cause our ladder issues, we did not see any bands from the p21 PCR (gel image not included).
Dylan prepared electrocompetent cells using the Myers and Myers protocol. Modified protocol using two 5mL cultures @ 4000g for 10 min. Washed with 2 mL sorbitol, resuspended in 100 uL sorbitol. First electroporation ts = 2.32 ms, second ts = 2.02 ms. Added 1 uL of plasmid (575.5 ng) to each cuvette. First used .60 V, then 0.55 V, both with R = 400 ohms.
Mark and Danielle started liquid cultures of S1, S9, S10, S11, S15, S16, S18, S27 (See: strain list)
July 3rd, Tuesday
Caleb miniprepped S16 (p15), S22 (p21), S9 (p8), S10 (p9), S11 (p10), S18 (p17), and S15 (p14). (See: strain list) Instead of a single EB elution, Caleb did two elutions, 30 ul each. The double 30 ul elution turned out to be effective at recovering a usable amount of DNA in the second elution, so we're including it on our miniprep protocol for future minipreps.
The rerun of our Gibson sequencing failed again, so we tried a more roundabout method of determining whether our 3 Gibson transformants have complete nah operons or not. We digested them each with BamHI, expecting specific fragment lengths on a gel electrophoresis.
When Dylan and Mark were attempting to determine what volume of ethidium bromide should be used in the gel, they came to the conclusion that better images may result from staining the gel in dilute ethidium bromide after running the gel, rather than including the stain in the gel. This experimental first staining used 200 mL of water conaining 20 uL of 10 mg/mL ethidium bromide, with gentle agitation for 1 hour. This stained the bands well, but also provided a large amount of background and took a very long time.
We only saw one band on each, all of them at about 3.6kb.
Because no transformations from our previous competent freezer stocks were successful, Dylan decided to make a starter culture of Shewanella strain JG 700 in preparation for making new competent stocks using the PNNL Protocol. Swati and Tina used this starter culture to complete the preparation.
July 4th, Wednesday
In the morning, Dylan, Swati, and Danielle prepared p8, p9, p10, p14, and p15 for sequencing (from the minipreps that Caleb preformed the previous day).
The team took the rest of the day off and had a barbeque at Buttermilk Falls. There was an excess of food, spontaneous song, and fireflies.
July 5th, Thursday
Upon arriving to the lab, Dylan dropped off the sequencing tubes we'd set up on the morning of the 4th for analysis. He also decided to do a Colony PCR of the potential transformant with the salicylate reporter plasmid. However, when setting up this reaction, Dylan realized that we'd used the wrong sequencing primers (we'd used the standard VF2 BioBrick primer instead of a custom primer for the pBBRBB backbone). Consequently, Mark and Dylan resubmitted p14 and p15 for sequencing while the colony PCR was ongoing. After visualizing the PCR products using DNA Gel Electrophoresis, Dylan concluded that the colony we'd screened did not have our plasmid of interest, and was the result of contamination.
Similarly, Dylan noticed small, evenly spaced colonies on all of the JG700 transformant plates from the Myers and Myers procedure we'd undertaken on the 2nd of July. Because none of the colonies looked red (as they should have if they were replicating pSB3C5), Dylan concluded that the chloramphenicol on the plates had degraded, and the colonies were resultant from untransformed cells.
Because the colony PCR of the potential salicylate reporter didn't yield good results, Dylan and Youjin set up another Phusion PCR to amplify the salicylate-sensing region from p21. Simultaneously, Mark made SOB media for recovery after future transformations, as we'd just received the ingredients for the broth.
Because we needed to submit for sequencing twice (because we used the wrong sequencing primers), Swati et al. set up liquid cultures of p21, p8, p9, p10, p17, and p14 in order to have more DNA in the freezer. We like having DNA in the freezer. (See: strain list)
July 6th, Friday
Caleb miniprepped p21, p8, p9, p10, p17 from the liquid cultures set up the previous night. (See: strain list) We couldn't miniprep p14, since – after checking notes from the previous night – we learned that the culture was made with the wrong antibiotic (chloramphenicol instead of kanamycin).
Some sequencing results came back, however the Gibson results are still confusing.
Also, a gel was run for the p21 PCR product.
July 7th, Saturday
July 8th-14th
The focus of this week was to transfer our arsenic sensing plasmids to Shewanella strain JG700 via congugation with E. coli strain WM3064. After struggling to electroporate control plasmids, we consulted with Dr. Gralnick who reccomended proceeding with conjugation and kindly provided E. coli strain WM3064. This strain is auxotrophic for 2,6-Diaminopimelic Acid (DAP), which makes the strain useful for selecting against non-Shewanella conjugants.
Another major decision this week was to give up on Gibson assembly - our sequencing results suggested that strange things had happened, and since we already had PCRed the full nah operon out of P. Putida, we decided proceeding with Gibson was not an efficient use of time. We were also unsure as to why p21's PCR was repeatedly failing, and decided to submit the part from the registry for sequencing, to try to troubleshoot what was going wrong.
July 8th, Sunday
July 9th, Monday
Dylan performed a Phusion PCR with a single annealing temperature of 59 degrees Celsius using primers 27 and 28 for the final time. In the future, new primers will be used. The purpose of this PCR was to amplify p21 (See: strain list). The PCR product was analyzed using agarose gel electrophoresis to determine if the desired 1.127 kb fragment is present. We talked to Dr. Jeff Gralnick about electroporating Shewanella, and his recommendation was to try conjugation instead. To transform our plasmids into Shewanella with conjugation, we had asked Dr. Jeff Gralnick for WM3064 E. coli for which plates containing 2,6-Diaminopimelic Acid (DAP) are required. In preparation to receive this strain, the plates with DAP and DAP/kanamycin were prepared.
July 10th, Tuesday
Caleb miniprepped strains containing Gibson products and the arsenic promotors without BamHI cutsites: plasmids p8, p9, p10, p15, and p16 (See: strain list) Also, our new freezer arrived. We cleaned and welcomed our new friend.The WM3064 came in and we plated from the agar stab. Dylan set up two 18 mL/hr continuous flow reactors using 10x diluted LB and innoculated one of the reactors with JG700 and one with wild type Shewanella oneidensis MR-1.
July 11th, Wednesday
Dylan started a WM3064 subculture in the morning before meeting Swati, Shweta, Caleb, and Danielle at the Boyce Thompson Institute for Plant Research to give a presentation both on our project and on the broader applications of synthetic biology to the energy industry to a group of high school teachers from New York state. After the presentation, Dylan and Danielle prepared plates containing DAP/chloramphenicol and DAP/ampicillin to grow transformed WM3064 on, while Mark continued preparing electrocompetent stocks of WM3064 with the subculture that Dylan had started.
We transformed things later that day.
July 12th, Thursday
Shweta prepared a glycerol stock of WM3064 for future use, and Dylan plated some remaining transformed WM3064 hoping to get more colonies of the successfully transformed cells.
Dylan and Mark submitted p8, p9, and p10 for sequencing using two different primers for each plasmid.
The previous night's p21 PCR seemed to have failed, even with the new primers. Based on the gel run by Claire, then analyzed by Dylan and Caleb.
July 13th, Friday
Today began with Dylan and Caleb setting up conjugations between our transformed donor E. coli and S. oneidensis.
The sequencing results for p8, p9 and p10 all came back as failed, so it was concluded that the Gibson assembly failed. There were five locations on the nah operon at which sequencing could begin, of which we considered all but the fourth. Sequencing at the first location resulted in a sequence that corresponded to the fifth sequencing location, with some sequence before it and all of the expected sequence after it. The fifth sequencing location produced the expected sequence. Then the middle two regions which we considered all resulted in failed sequencing. This has led us to the conclusion that segments that are required in the operon are missing, and thus the assembly failed. Dylan and Caleb began attempting to look into the possibility of this being a cause, and some potential solutions.
In discussing why p21 PCR seems to continue failing, the idea of having p21 sequenced arose. Dylan prepared the sample and submitted it for sequencing. Meanwhile, Mark set up yet another Phusion PCR of p21, this time with a higher annealing temperature of 61 degrees Celsius and a decreased extension time of 18 seconds. The goal was to prevent the mispriming which seemed to have occurred in the previous day's attempt.
Later that day, Dylan set up yet another PCR with a decreased annealing temperature of 55 degrees Celsius in order to empirically narrow-in on the optimal annealing temperature for the amplification of the salicylate sensing region from p21.
July 14th, Saturday
Swati ran a gel of 5uL of the p21 PCR that Mark set up yesterday. After staining, the DNA ladder was well visualized but there were no bands in the p21 lane, suggesting that the PCR did not work. In a fit of rage, Swati yelled at Maneesh for going to get a drink of water - yet another innocent victim of failed PCR. Then, after a full team meeting, we all came together for a dumpling party. Noodles, dumplings, and gnocchi of all kinds were eaten by the voracious team. In the spirit of true, unwavering scientific inquiry, we conducted a novel experiment whilst feasting on these delectable treats. The result: fried Dorito dumplings are delicious.
July 15th-21st
July 15th, Sunday
Dylan noticed that we had colonies on the plates re-streaked from the original plates of conjugated S. oneidensis. He started overnight cultures of S20 (See: strain list) in kanamycin so that we can sequence and make glycerol stocks. He also started overnight cultures of w.t. S. oneidensis to innoculate into new reactors in Riley Robb, which will be used as positive controls.
July 16th, Monday
The overnight cultures of S. oneidensis need more time to grow - overnight cultures of these leisurely bacteria should be started at noon the previous day, instead of in the evening as with speedy E. coli. Dylan ran another Phusion PCR of the entire nah operon, since we got a lot of mispriming the first time we ran it. The thought was that we had run the previous PCR at the optimal temperature for Vent but used Phusion, so Dylan set up the new PCR correctly, with a single annealing temperature of 66 degrees Celsius and a lengthened final extension time of 15 minutes to account for the size of the nah operon (~10kb). Caleb ran a gel of the PCR and visualized a single band ~9.5kb. PCR of the nah operon out of p20, P. putida (See: strain list) was successful! Tina and Swati gel extracted and quantified, extracting two samples at 38.8 ng/uL and 27.2 ng/uL.
Spencer miniprepped p14, our arsenic reporter part (arsR + mtrB w/ BamHI cutsite), from S20 (See: strain list) for sequencing, recording yields of 44.3ng/uL & 35.8ng/uL (colony 1) and 50.6ng/uL & 53.5ng/uL (colony 2) from quantification.
Dylan and Tina set up transformations of p15 and p16 (See: strain list), the arsenic reporter part without a BamHI cutsite, so that we can conjugate S. oneidensis in the next few days. They also set up a transformation of the miniprepped BBa_J01003 with the oriT mobility gene. Our new method of inserting plasmids into S. oneidensis, using conjugation rather than electroporation, requires that all our plasmids have the mobility gene. However, the plasmid we were going to use for the nah operon (pSB3C5) has no mobility gene. Possible approaches to take are to clone oriT from iGEM kit plates into pSB3C5, or to PCR the mobility gene out of one of our own plasmids and clone that into pSB3C5. We will try to get oriT from kit plates first.
July 17th, Tuesday
Shweta set up a Phusion PCR of p21 (See: strain list) to try again to extract the salicylate-sensitive promoter. Sequencing of p21 showed that there was a stem loop sitting upstream of the ENX biobrick cutsites, which wasn't expected and is most likely why our primers are not effective. Before redesigning primers, we are going to try PCR with the standard iGEM forward sequencing primer and the same reverse primer that we designed for p21.
July 18th, Wednesday
Dylan set up p15k, p16k conjugation plates from overnight cultures of transformed WM3064 and JG700. Incubated 8 hours, streaked for single colonies on kan plates with E.coli and Shewanella controls. Caleb miniprepped p24a (BioBrick part with oriT) from overnight culture. Desalted overnight ligation mix and transformed DH5α (nah operon in pSB1C3).
Restreaked... p26a p30a p27a p29a
Set up reactor in Riley Robb.
Spencer made overnight cultures of p25a, p28a, p31c.
July 19th, Thursday
Only observed 2 possible colonies from transformants from yesterday's ligation (nah operon in pSB1C3). Restreaked from one of these colonies along with a DH5α control.
We got single colonies on the JG700+p15 and JG700+p16 plates, and no growth on control plates. Tentative conclusion is that conjugation was successful. Made reference plates of picked colonies, and grew up liquid cultures from same colonies for sequencing.
Dylan and Caleb miniprepped p25a, p28a, p31c from overnight cultures, then made glycerol stocks of S25, S28, and S31.
Inoculated reactor in Riley Robb with S20, our engineered arsenic reporter strain.
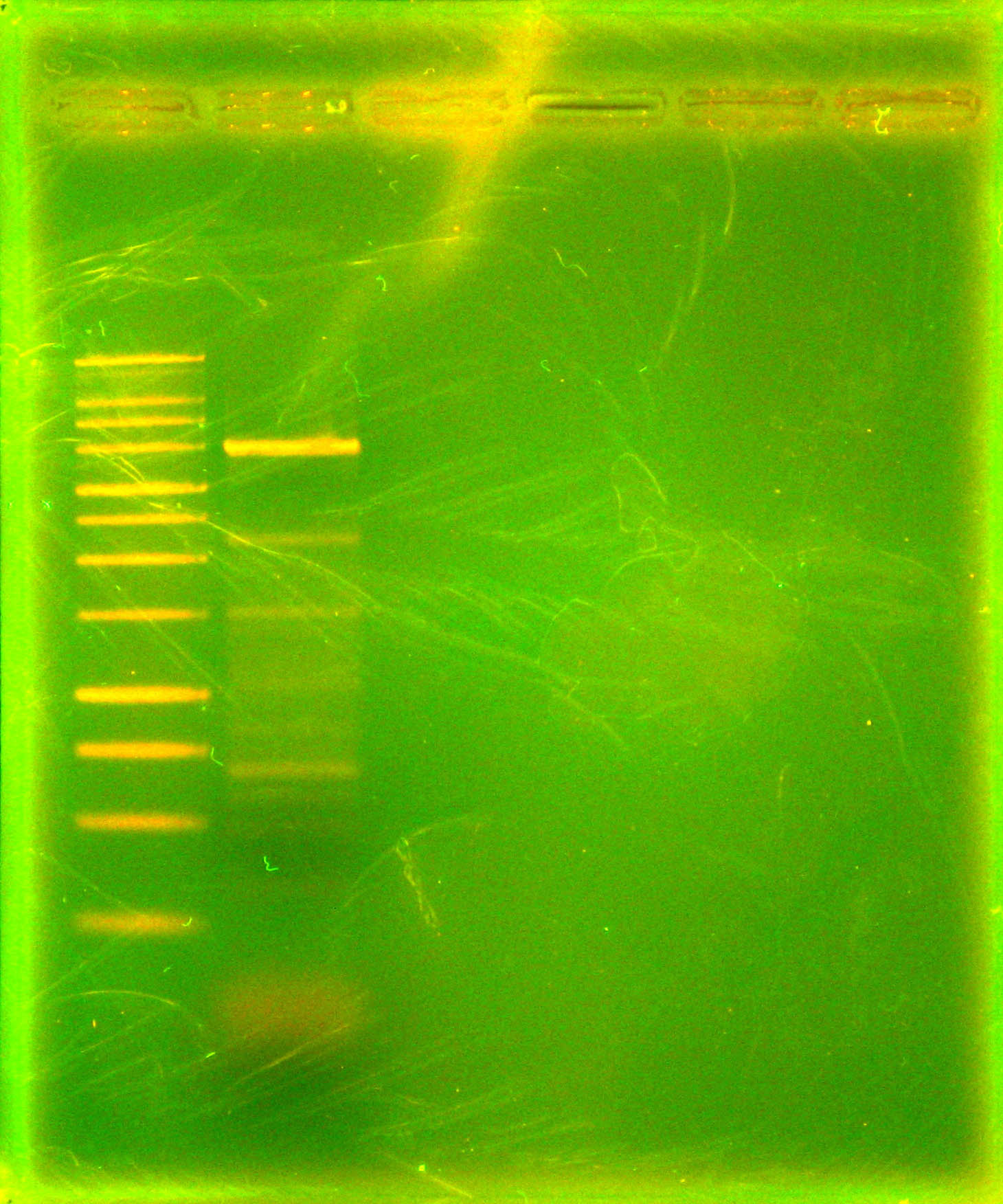
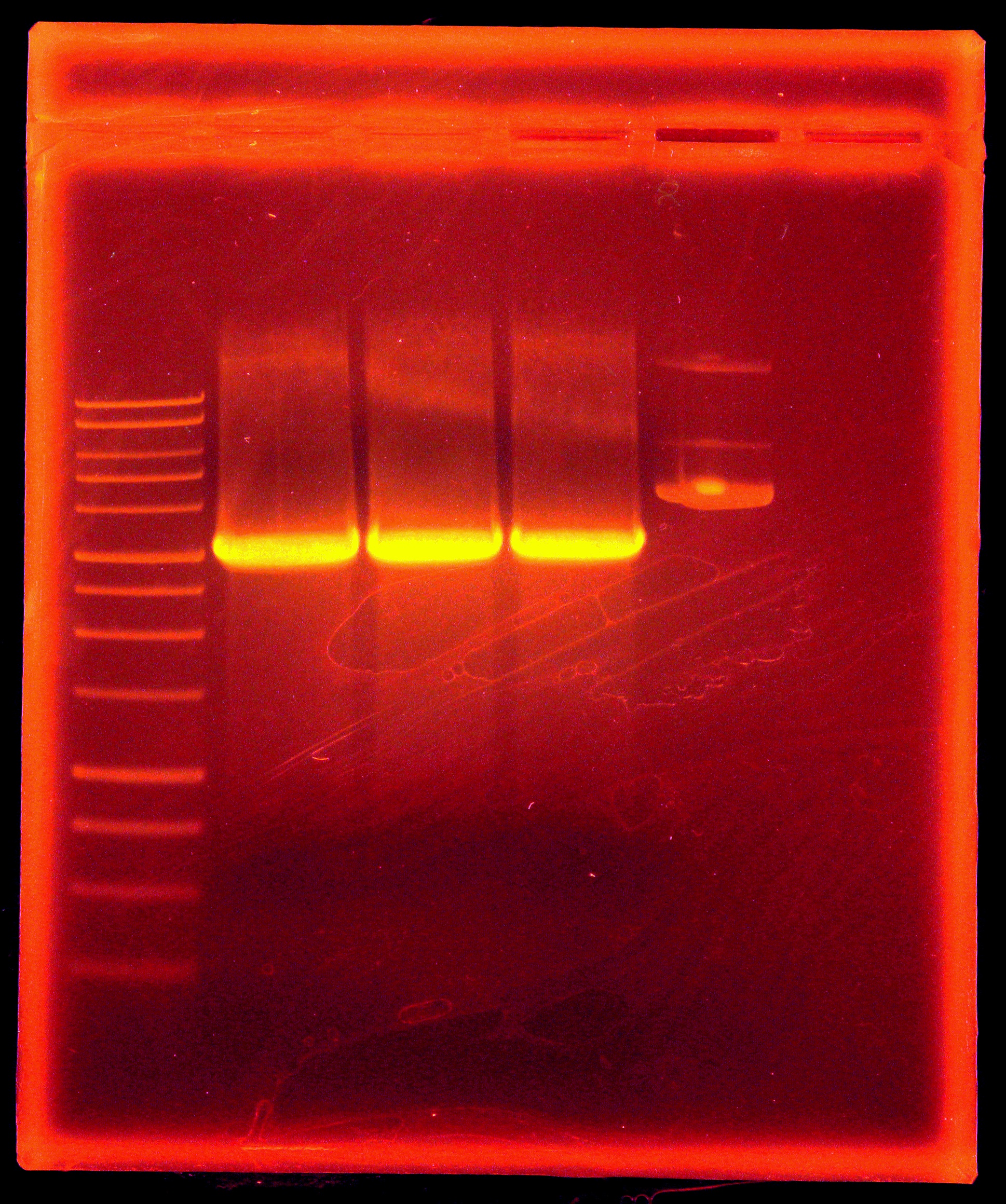
Digested p21 PCR product along with miniprepped p14 with EcoRI and AscI. Purified p21 PCR digestion with Omega Bio-Tek E.Z.N.A. MicroElute DNA Clean-Up Kit. Dephosphorylated p14 digestion with Antarctic Phosphatase, and ran entire mixture on a gel. (Picture of gel). Purified 6kb band with Qiagen gel extraction kit.
Overnight cultures of a bunch of stuff.
July 20th, Friday
Shweta and Tina miniprepped p26a, p27a, p29a, p30a (Anderson series constitutive promoters), and p14k from overnight cultures.
Dylan set up 30 minute room temperature ligation to construct salicylate reporter with digested p21 PCR product and p14 (isolated yesterday, quantified today). Not confident that ligation will work because of incredibly low yields from digestion cleanup and gel extractions.
Because Shewanella grows more slowly than E.coli, Claire did a second miniprep of p15k and p16k from JG700 in order to submit plasmid for sequencing to confirm that conjugation was successful.
In anticipation of the failure of Dylan's ligation, Swati set up 3 Phusion PCR reactions in parallel, each with the same template (p21), repeating the parameters that had proven successful (no mispriming) previously.
Dylan set up overnight cultures of... nah operon in pSB1C3(?) S18 (DH5α + pSB3C5, p17c) WM3064+p14
July 21st, Saturday
July 22nd-28th
The goal of this week was to transform DH5α with nah operon (p20 PCR + p25a, p27a, or p29a).
July 22nd, Sunday
Danielle and Dylan digested p14(Arsenic reporter) and p21(Salicylate sensing region) for ligation. Swati ran a Phusion PCR to amplify p21. Then Dylan ran a gel electrophoresis and did gel extraction to isolate p21 PCR digest and p14 backbone.
July 23rd, Monday
Dylan quantified the gel extractions, quite successfully with a newly invented protocol. Then he dephosphorylated p14, and submitted some DNA for sequencing. At one point, the Magical Graduated Cylinder of Elmira (500 mL) fell from the sky and shattered in the sink. Some voodoo was clearly in the air. It appears that the ligation products from the day before were either not created or not successfully transformed, as the plates contained no colonies.
Caleb and Tina prepared more kanamycin and chloramphenicol plates while Mark desalted Dylan's ligation product. Then Dylan, Mark, and Tina transformed DH5α with the ligation product, p33k, p14k, and p31c (See: strain list).
July 24th, Tuesday
In the morning, Dylan did Phusion PCR of p20(nah operon). Then Caleb and Dylan did a double digestion of p20 PCR and p17c for ligation. Dylan ran a DNA gel electrophoresis to confirm that the previous PCR was successful. Steven performed gel extraction to isolate p17c backbone fragment.
July 25th, Wednesday
Mark set up two ligations of a p20 PCR product and p17c, one at standard concentrations and one at very high concentrations. (See: strain list) p20 PCR was cut at Xbio1 and Spe1, which have compatible sticky ends. This means that p20 PCR can be ligated into backbone in two directions, only one of which is useful. These were desalted and transformed by Mark and Dylan, along with p33k. Meanwhile, Tina and Chie set up double digestions of p24a, p20 PCR product, and single digestions of p25a, p27a, and p29a.
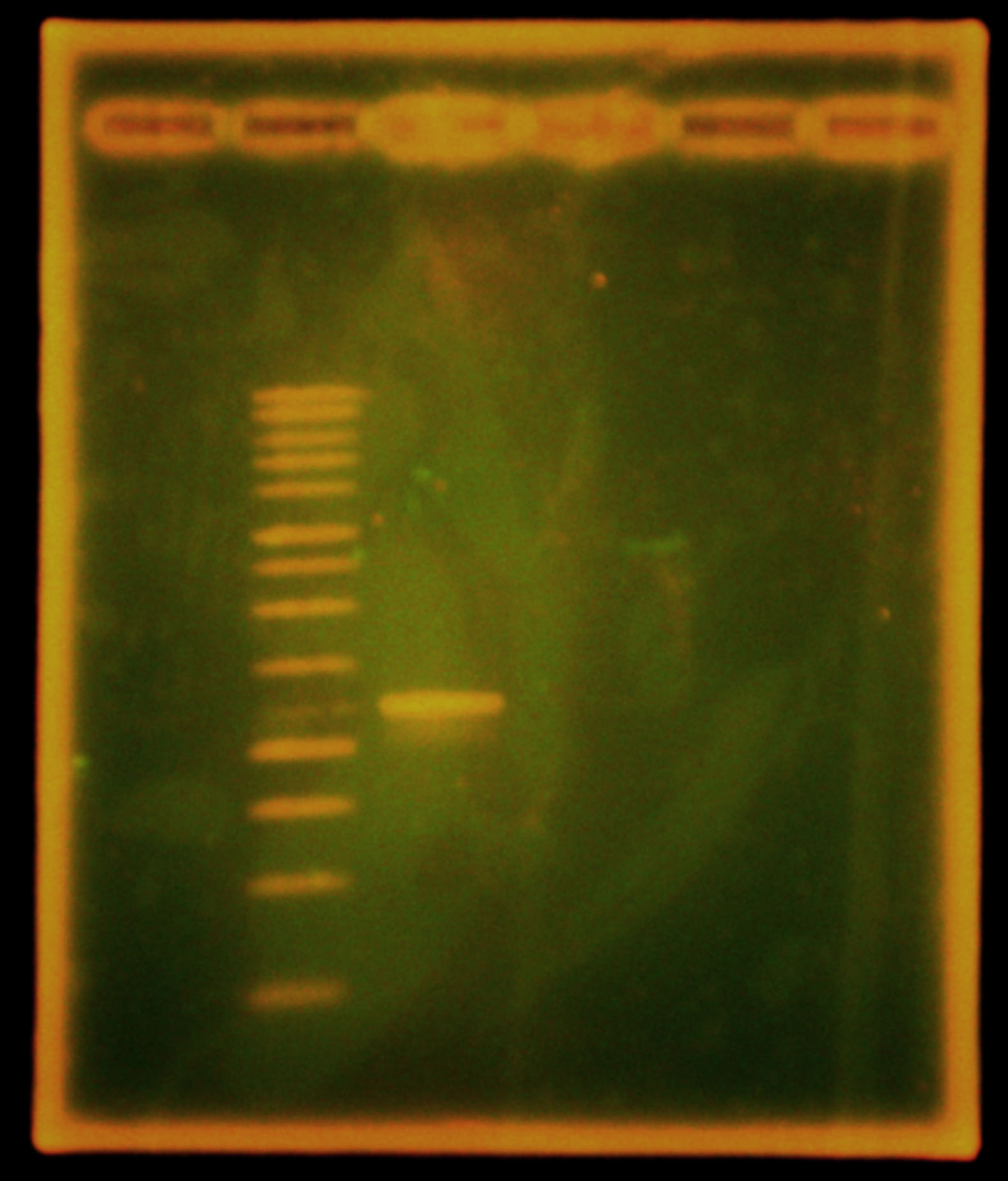
Dylan ran two gels of the double and single digestions. The single digestions of the Anderson series promoters showed single bands, which ran slightly faster than the supercoiled DNA control. This may be because the supercoiled DNA was nicked. The gel of the oriT double digestion showed more long bands than expected – possibly a result of star activity – but had a band ~400bp which was interpreted to be the insert of interest.
July 26th, Thursday
Caleb gel purified the digestion products of p24a, p20 PCR, p25a, p27a, and p29a. Danielle dephosphorylated products of p25a, p27a and p29a in preparation for ligation.
July 27th, Friday
In the morning, Claire miniprepped p11k, p17c, and p33k from the overnight cultures that Dylan had set up. Meanwhile, Dylan heat killed the ligase from the overnight ligations(p25a, p27a and p29a), while Caleb prepared to desalt the ligation mixtures before electroporation via drop dialysis. Shortly thereafter, Dylan and Caleb electroporated the four desalted mixtures into DH5α, allowing the cells to recover one hour in SOC before plating. If these transformations are successful, we will have cells that carry plasmids expressing the nah operon under the control of three different constitutive promoters with varying strength, as well as a cell line that carries a plasmid that confers chloramphenicol resistance, has a p15a origin of replication, and an origin of transfer. Eventually, the nah operon will be ligated as an insert into such a backbone.
In the evening, Dylan and Tina started 30 mL overnight cultures of S24, S25, S27, S29, S15, and S17 to miniprep from the next morning. S24 carries a plasmid with an oriT, which will be miniprepped in case our previous ligation of an oriT into p17c failed. S25,27, and 29 all carry plasmids with Anderson series constitutive promoters of varying strength, which will be used to set up fluorescent controls to monitor mtrB expression levels. S15 and S17 carry our engineered arsenic reporter plasmids, which will be further modified to facilitate control studies in mtrB expression.
After setting up the cultures, Dylan and Swati left Weill and headed over to Riley Robb where they assembled three new reactor setups in the Angenent lab. Two of these reactors will be run in continuous flow operation with a BioLogic potentiostat, while the other will be run in batch mode with a CH Instruments potentiostat. Tomorrow, one continuous flow and one batch reactor will be inoculated with wildtype Shewanella oneidensis MR-1, while the remaining continuous flow reactor will be inoculated with our engineered arsenic reporter strain, S20. The purpose of running the continuous flow reactors is to better define the maximum current output we can expect from our induced strains in response to analyte, and to repeat an experiment carried out on the CH potentiostat for basal activity from the uninduced arsenic reporter. Because we were unsure of the veracity of the data from our previous experiment using the CH potentiostat, we are running the batch reactor to see if the observed current response is what we'd expect from wildtype Shewanella.
July 28th, Saturday
Today, Swati did six minipreps in the morning and it took her four hours...and it paid off! The nanodrop told us that our highest yield was 812.6 ng/uL.
While Swati was devoting her soul to the E.Z.N.A kit, Dylan continued to set up reactors in the Angenent Lab, making reference electrodes for the three reactors, and getting the pumps set up for the continuous flow reactors. After our weekly team meeting, he inoculated the reactor.
Swati prepared overnight cultures and reference plates, while Danielle prepared kanamycin plates and did autoclave.
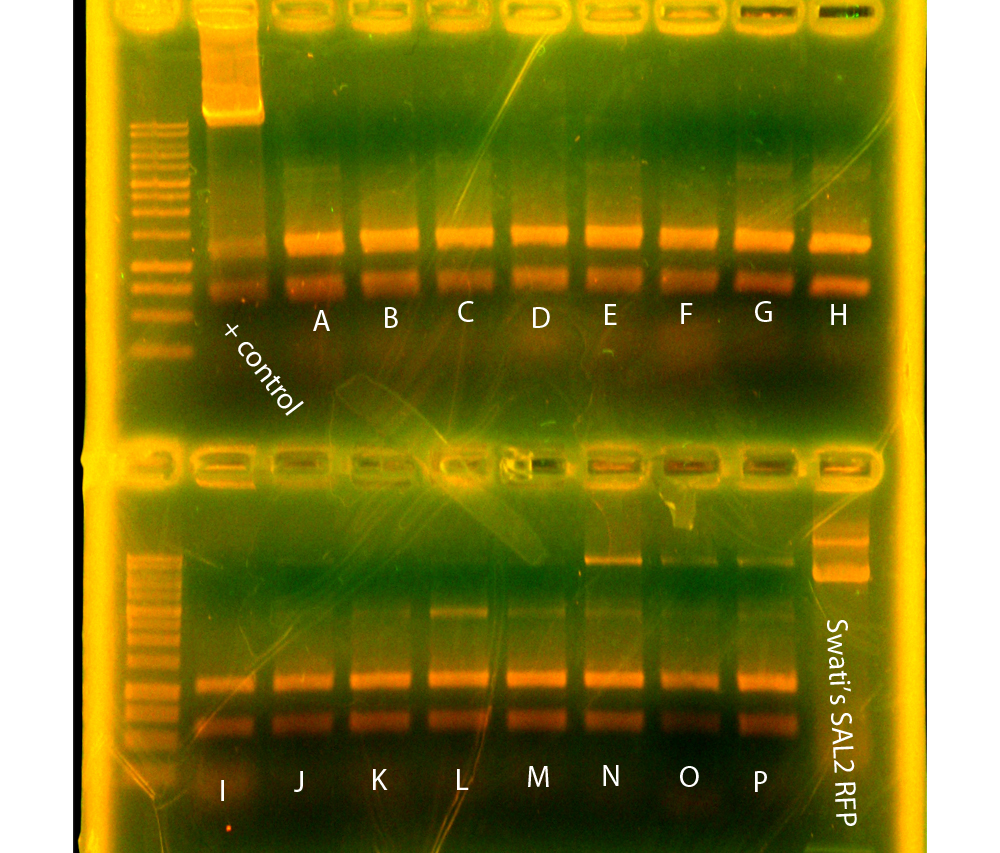
In the evening, Dylan and Swati did digestions for RFP controls and SAL2 reporter. Also, SAL reporter was transformed into WM3064.
July 29th-31st
SAL (the version of the salicylate reporter with the BamHI cutsite) was conjugated into JG700, and we created SAL2 (salicylate reporter without BamHI cutsite). We prepared constitutive controls and our reporters for fluorescence testing. We also continued cloning work to get the nah operon with a constitutive promoter ready to conjugate into JG700.
July 29th, Sunday
Apparently, Swati has yet to get over her miniprep craze. While most of the wetlab hiked through Danby State Forest, Swati did more minipreps: p25a, oriT + PSB3C5, and nah operon + p29a. The cultures of S25 and nah p29 were bright magenta, while oriT in PSB3C5 was a pale tan. However, the last of these contained an unidentified crystalline substance, possibly a byproduct of producing the nah operon. Two other cell cultures also didn't grow, which was disheartening, as it meant there were less minipreps for Swati to do. Interspersed between the copious miniprepping were two gels to purify the digestions from the previous evening, all of which appeared to work! Dylan and Claire, returning from their nature trek, joined Swati in lab, finding Shweta and Eric along the way. While Dylan took over Swati's gel extraction, Swati, Shweta, and Claire discussed upcoming outreach events. As Ecologist Claire explained the feeding habits of Tomiculus troutus (AKA Tommy the trout), Eric began to feel left out and, in the process of showing off the website, discovered Dylan's odd love for typography. After spending roughly an hour accomplishing nothing, Dylan and Swati set up liquid cultures in preparation to put SAL into JG700. Dylan returned to his reactor-ridden lair for a short spell.
July 30th, Monday
In the morning, Dylan set up conjugations between WM3064+SAL and JG700 while Caleb quantified the minipreps and gel extractions from Sunday. After quantification, he submitted nah+p29a and oriT+p17c for sequencing. Dylan and Mark dephosphorylated the digestions of SAL with SpeI & PstI, p14k with SpeI & PstI, p16k white SpeI & PstI, p16k with EcoRI & AscI, and p11k with EcoRI & PstI. Tina and Danielle then set up ligations of mRFP+SAL, mRFP+p14, and mRFP+p16 to put mRFP downstream of the arsenic and salicylate reporters, in order to be able to characterize our induced promoter strengths by measuring fluorescence. We also ligated p16+SAL to make SAL2, the salicylate reporter without the BamHI cutsite. Finally, we ligated p25, p27, and p29 (the Anderson promoters with mRFP) into pBBRBB, in order to be able to put them into Shewanella.
July 31st, Tuesday
Mark made DH5α electrocompetent stocks. Dylan checked conjugation plates to put SAL into JG700. He then made overnight cultures of JG700+SAL and S31 (PSB1C3, as well as WM3064 to prepare competent stocks. Dylan autoclaved a reactor in Riley-Robb and set it up in batch with M4 media, and inoculated with wild-type S. oneidensis MR-1. The purpose was to repeat a previous batch experiment in order to be sure that it is something about feeding with LB that is decreasing current production.
Sequencing results came back on nah+p29 and oriT+p17c and things looked good. Consequently, Dylan decided to digest nah+p29 with EcoRI and SpeI, and oriT+p17c with EcoRI and XbaI in order to eventually ligate the nah operon with a constitutive promoter into the p17c backbone, upstream from the oriT sequence (in order to allow the plasmid to be conjugated into Shewanella). Claire prepared these digestions, along with digestions of p31c and p20 PCR product (in order to get the nah operon into pSB1C3).
Dylan and Caleb desalted the previous night's ligations and than transformed into WM3064, since we will want to get plasmid into Shewanella.
August
August 1st-4th
August 1st, Wednesday
In the morning, Dylan noted that 5 of the 7 transformations from Tuesday produced plates with colonies. Those that worked were three Anderson series constitutive promoters (with downstream mRFP) in pBBRBB and the versions of our two arsenic reporters with mRFP downstream of mtrB. In the evening, Swati and Dylan prepared overnight cultures from all transformants and JG700, both to set up conjugations to get plasmid into Shewanella and to screen isolated plasmids for successful ligation. We will use these constructs to characterize the activity of the arsenic-sensitive promoter in Shewanella with respect to known constitutive promoter strengths. The two transformations that failed were versions of the salicylate reporter.
After staring at plates, Dylan ran the nah+p29, oriT+p17c, p31c, and nah operon (from p20) digestions on a gel, extracted the fragments of interest, and then dephosphorylated the backbone. The isolated digestion products were used later used by Steven, who set up two overnight ligations – one to put the nah operon, with a constitutive promoter, on a pSB3C5 backbone with an added oriT sequence, and one to put the nah operon into the MCS of pSB1C3 for subsequent site directed mutagenesis.
While Dylan ran his gel, Caleb miniprepped from the JG700+SAL and S31 cultures. After quantification, SAL plasmid isolated from JG700 was submitted for sequencing in order to confirm successful conjugation.
Meanwhile, Mark decided that he enjoyed making DH5a electrocompetent stocks on Tuesday so much that he had to make more WM3064 electrocompetent stocks today. Starting from subcultures that Dylan had prepared, he prepared the stocks, and was assisted by Danielle.
August 2nd, Thursday
Caleb and Dylan created conjugation plates of p14RFP, p16RFP, and p25-29BB as well as miniprepped from the same plasmids in WM3064 (see: strain list). Dylan, Mark, and Tina then desalted and transformed SAL2 and SALRFP into WM3064 as well as p31c_nah and oriT_nah_p17c into DH5a. Tina and Dylan then quantified the minipreps from earlier in the day. Dylan also digested pBBRBB and p33k with EcoRI & PstI to put lac inducible mtrB into the pBBRBB backbone.
August 3rd, Friday
After running a digestion of p33k, the lac inducible mtrB part from the parts registry, we discovered that the part does not include mtrB! We should have seen a ~3.6kb band on our gel after digesting with EcoRI and PstI, but instead isolated a band ~1.5kb. After checking the sequencing on the parts registry we discovered this was because the part didn't include mtrB, so we won't be able to use it as a positive control for inducible mtrB.
Sequencing of p37k-p41k from WM3064 came back good, meaning we can put these control parts, with RFP downstream of our various reporters, into Shewanella via conjugation. Hence, conjugation was carried out.
August 4th, Saturday
August 5th-11th
August 5th, Sunday
Swati yet again was saddled with a massive number of minipreps, since her magic fingers are able to cast yield-increasing spells on minipreps. She miniprepped: SAL2 from WM3064, p37-41k from JG700 (one arsenic reporter with cut sites flanking the RBS, another arsenic reporter without the cut sites, and three different Anderson series promoters with mRFP1 downstream on a pBBRBB backbone), oriT p29nah_p17c from DH5a, and nah_p31c from DH5a. And indeed her yields were impressive, massive, gargantuan! Good job Sorceress Swati.
We will sequence SAL2 and oriT p29nah_p17c to see if we should continue to conjugation, and sequence nah_p31c to see if we can start site-directed mutagenesis to get a biobrick-compatible nah operon on the biobrick backbone. We will sequence or PCR to confirm p37k-p41k's conjugation into Shewanella.
August 6th, Monday
Caleb and Dylan amplified the two different arsenic promoters (p37k and p38k) and ran them on a gel alongside p14 and p16 (the arsenic reporters without mRFP1 downstream) to confirm successful insertion of RFP downstream of mtrB in our arsenic reporter parts in Shewenella. Band lengths appeared in expected places. Dylan is worried that we will not be able to use PCR to definitely confirm that p39-41k worked, so we submitted these for sequencing. Unfortunately, sequencing failed, perhaps because random gunk from Shewanella added noise to the process.
Dylan also ran a gel of nah operon PCRs to make sure that there was no mispriming. The gel looked good, so he submitted the PCRs for sequencing.
August 7th, Tuesday
JG1531 overnight culture didn't grow. We suspect that the plate Dylan picked from is dead. We'll have to go back to the glycerol stocks if we want to play with mtrE. Dylan set up a continuous flow M4 reactor in morning. Caleb started a liquid culture of MR-1 with which to inoculate the reactor. Checked sequencing results of nah stuff. oriT thing was bad. nah_p31c was good.
Caleb and Dylan proceeded with site directed mutagenesis of nah_p31c, attempting to get rid of the first PstI cutsite in the nah operon. After digestion with DpnI, we purified using the E.Z.N.A. MicroElute kit and quantified. DNA was split into three directions: First, Danielle and Dylan set up another mutagenic PCR using the digestion product as template to get rid of the second internal cut site (we expect lower mutation efficiency because template DNA is not methylated). Second, we transformed DH5a with the mutated plasmid. Third, Danielle and Chie digested both the purified – and hopefully mutated – plasmid and un-mutated plasmid with PstI. Dylan ran these digestions on a gel, along with supercoiled plasmid as a control.
In the evening, we didn't observe growth in the MR-1 culture, so Dylan set up another culture just in case Shewanella was dead and not lazy.
August 8th, Wednesday
Caleb and Dylan performed another digest to check if the second site-directed mutagenesis (that was supposed to get rid of the second PstI internal cut site within the nah operon) worked. Unfortunately, there wasn't enough DNA to see anything when visualized on a gel. They decided to proceed with electroporation into DH5a just in case - however, to use Caleb's terminology, cells exploded - there were probably too many salts in the solution, which causing arcing and a PBBHTTTZZZ! of cells all over the cuvette.
Dylan re-digested p29 nah and oriT p17c to redo a ligation that would allow for conjugation into Shewanella later on. Dylan also re-digested p27 and SAL to redo a ligation to create a plasmid with SAL-RFP.
Dylan also submitted several samples for sequencing to make sure that three Anderson promoters with RFP downstream in JG700 and SAL2 (the salicylate reporter without the BAMHI cut site) in WN3064 and JG700 were in the correct sequence.
August 9th, Thursday
Today Dylan chilled with his mom. Well, he tried to. :) Despite the eternal bonds that bind mother and son, plus the 3 billion miles she flew to see him, he still couldn't resist coming in to lab to open packages! And then got sucked into a long discussion of what had to be done for the rest of the day - scientific endeavors taking him away yet again from filial duties. Swati and Claire bonded over dry ice, and Caleb regaled us with tales of dry ice bombs gone awry.
Swati and Caleb miniprepped the first mutagenesis of nah_p31c that had been transformed into DH5a yesterday. They then digested it with PstI-HF to see if the mutation was successful. Unfortunately the mutagenesis failed! We will start over from nah_p31c and lowering the annealing temperature at 60degC. The Stratagene kit, which uses PFU ultra, asks for 60degC, but because we are using our own boot-leg protocol with Phusion we did the first try at 65degC, as Phusion usually calls for a higher annealing temperature than the theoretically calculated value. For this second try we will stick to the 60degC suggested by Stratagene and see if we get better results. Also, called NEB to find out if after PCR with Phusion, DpnI will still have activity in the following digestion step in Buffer 4, or if we need to clean up the PCR before digesting with DpnI - they said PCR clean up isn't needed.
We also ran digestions for making the nah operon on a backbone with a mobility gene and the salicylate reporter (w/ BamHI cutsite) with RFP downstream. We cut our mobile backbone, OriT in p17c (pSB3C5), with EcoRI and XbaI, while cutting the nah operon (p29nah) with EcoRI and SpeI. The nah operon with mobility gene must be constructed so that we may conjugate into Shewy and start testing our salicylate reporter. The salicylate reporter and p27a (from the Anderson series), with RFP, were cut with SpeI and PstI. The salicylate/RFP part will be used for troubleshooting the salicylate reporter. Digestion products were run on a gel, extracted, and quantified. Swati then dephosphorylated backbones and ligated both parts.
Sequencing for p39-41k didn't look good, and neither did the salicylate reporter w/BamHI cutsite miniprepped from JG700. The sequencing for salicylate reporter w/ BamHI in WM3064, however, looked good, suggesting that conjugation may not have been as efficient as we had hoped. After a pow-wow we decided not to sequence more colonies, as we are hoping some may be good, and more importantly that if we use qPCR to get quantitative characterization data, we won't need to use the RFP parts. The plates will stay in the fridge as a back-up plan.
In other news: Claire cried because it was difficult to update the notebook with a week's worth of work. Mark's dedication to the notebook is laudable and impressive. Good job team for doing so much! My head can't even comprehend the magnitude of your endeavors.
August 10th, Friday
Caleb miniprepped a plasmid with mtrE from JG1531, and just for kicks, miniprepped from the "exploded cells" from August 8th. Suprisingly, he ended up getting decent yields for both, showing that the electroporation worked despite arcing. Caleb then digested the nah operon of the miniprep with PSTI and NotI to check if the mutagenesis was a success. It was run on a gel alongside a PCR of the Anderson series promoters with RFP downstream and SAL2 from Shewanella (to check if SAL2 and the promoters were sucessfully conjugated after sequencing on Monday failed). Unfortunately, bands did not appear where we expected them to.
Steven and Spencer performed a PCR to get mtrE out of the Gralnick (JG700) plasmid.
August 11th, Saturday
Spencer and Steven checked the nah operon PstI and NotI digest (because yesterday's gel ran weirdly) and their mtrE PCR from the previous day on a gel. Unfortunately, bands did not appear where we expected them to.
August 12th-18th
August 12th, Sunday
Weekend overview: It looks like the mtrE primers are actually working to amplify the part out of JG1531. We'll be putting mtrE in p31c, both so that we may submit the novel part to the registry, and to begin site-directed mutagenesis.
We'll use mtrE mutagenesis as something of a control for nah operon mutagenesis (to see if the large size of the plasmid and nah operon is the problem).
It also looks like nah operon mutagenesis hasn't worked. We'll put this on hold, and continue with ligation of p29nah into oriT_p17c once desalting paper arrives.
Also, we'll have to redo things to confirm p39-41, SAL2 in JG700. What was done over the weekend looks weird.
August 13th, Monday
Dylan ran four simultaneous Phusion PCRs with an annealing temperature of 67degC and an extension time of 45sec to amplify mtrE. The gel looked good, with no mispriming and one band ~2.2kb. Claire did PCR clean up of the product, which will eventually be cut and ligated into p31c (see: strain list).
Because the results from the weekend's screenings were confusing, we also ran PCRs of p39-41k (miniprepped from JG700 strains) to confirm whether conjugation into Shewanella was successful. These plasmids contain mRFP under the control of constitutive promoters of varying strengths. After confirmation, we will use these parts to characterize our inducible promoters in Shewanella.
Dylan also started a liquid culture of JG700 + SAL to be inoculated into a continuous flow reactor with M4 media. With this setup, we will begin characterizing our reporter strains, initially in response to salicylate.
August 14th, Tuesday
August 15th, Wednesday
This morning Dylan noted that we may be getting a detectable basal level of current - around 6mA - in a continuous flow setup. This would make it possible to bypass the addition of mtrE to the salicylate sensing part, and still be able to distinguish Shewanella not detecting salicylate from dead Shewanella. He then added salicylate to the reactor innoculated with our salicylate reporting strain, bringing the concentration to 10uM. Later in the day, he noted that the current had risen to around 9mA, which is promising: our strain may be working! Claire set up four hour digestions with XbaI and PstI to put our arsenic parts, p14 and p16, into the iGEM backbone for submission.
Dylan also transformed a salicylate reporter with mRFP downstream into DH5alpha and WM3064, as well as the nah operon with the p29 promoter in a p17c backbone with an oriT. The second of these could be conjugated into Shewanella if transformed successfully. The first, the salicylate reporter with mRFP downstream, is a fluorescent version of the salicylate reporter. The fluorescent versions of our reporters is an alternative to qPCR to measure the relative expression level of mtrB: if we know what mRFP under the control of a constitutive promotor in Shewanella looks like, we can add arsenic or naphthalene/salicylate to our fluorescent reporter parts until reaching the same level of expression. Then, we can correlate this concentration of arsenic or salicylate to a certain strength of induced expression.
In case the fluorescent SAL reporter was not successfully ligated, we are preparing more mRFP, SAL, and SAL2: - In the morning Claire also cleaned up the digestions of SAL, SAL2 and mtrE PCR. However, due to a silly mistake on her part involving wash buffer and absolute ethanol (absolutely missing, to be precise) we will redo the digestions in order to get more DNA for putting these reporters into the iGEM backbone, and to put mRFP downstream. She then set up four hour digestions with XbaI and PstI to put our arsenic parts, p14 and p16, into the iGEM backbone for submission. - We digested mRFP with SpeI and PstI to make the part with mRFP downstream of mtrB in the salicylate reporters. We have already successfully put mRFP downstream of mtrB in our arsenic reporters, but it did not seem to work in the arsenic reporter. The gel for mRFP looked as expected, with the band for the insert slightly shorter than 900bp. However, on the same gel the the mutagenesis trial of the nah operon, digested with PstI and NotI, showed only one band. If mutagenesis had not worked, we would expect more than two bands, and if it had worked we would still expect two bands. Therefore we are unsure how to interpret this result, which we have seen twice now, but are going to start over with site-directed mutagenesis of the nah operon. We will also visualize the unmutated nah operon, digested with PstI and NotI, on a gel to see if that looks like we expect it to.
Finally, it should be noted with jubilance that Claire was reunited with her umbrella today! The joy in the lab at this event was palpable and will be remembered for years to come.
August 16th, Thursday
Today, we continued work to get our engineered reporters into pSB1C3 for submission to the parts registry. In the morning, Dylan miniprepped both versions of our salicylate reporters, along with more pSB1C3 from overnight cultures of the corresponding DH5a strains. Following quantification, he set up digestions of each miniprep with XbaI and PstI. (We decided to cut with XbaI because we discovered an extra base pair between the NotI and XbaI cutsites in the normal BioBrick prefix, an artifact of a previous team's work). Caleb also dephosphorylated previously isolated pSB1C3 backbone to prevent self-ligation, while Steven gel extracted the salicylate reporter inserts and more pSB1C3 backbone (to be quantified Friday morning).
August 17th, Friday
August 18th, Saturday
August 19th-25th
August 19th, Sunday
Summary goes here.Click to view the details.
Because all other transformations seemed to work, we grew up overnight cultures from single colonies on each plate. We will miniprep from these cultures and screen the minipreps for the correct insert. If confirmed, we will have successfully have put both versions of our arsenic reporters, as well as both versions of our salicylate reporters inside p31c (i.e., pSB1C3), as is required for registry submission. We will also have successfully put mRFP downstream of mtrB on SAL2, which will be used for characterization of the salicylate sensitive promoter in S. oneidensis.
August 19th, Sunday
August 20th, Monday
August 21st, Tuesday
August 22nd, Wednesday
Conjugating nah into Shewy: Caleb, Tina, and a guest Louis miniprepped 5 plasmids containing nah operons with constituitive promoters from a WM3064 plate and miniprepped one plasmid that we previously confirmed as having the nah operon (without a promoter) to use as a positive control in the future.
August 23rd, Thursday
August 24th, Friday
Conjugating nah into Shewy:
We wanted to accomplish two things:
- Amplify the nah operon with a promoter using primers with appended cut sites.
- Optimize colony PCR for future Cornell iGEMers (past attempts have failed.)
To accomplish both of these objectives, we did Phusion PCRs using plasmids containing nah operons with constituitive promoters as a template and a Phusion colony PCR using the same colonies we miniprepped the plasmid from. Because we expect the miniprep PCRs to work, doing the colony PCRs in parallel will allow us to determine whether our colony PCR technique is working or not. For our colony PCR purposes, we want to be able to screen for colonies while being certain we can go back and miniprep the same PCRed colony. Our colony PCR method is as follows:
NOTE: We later determined there was a cataclysmic flaw in the following method, can you figure it out? (wink wink nudge nudge...)
- Label colonies of interest and correspondingly label microfuge tubes
- Add 50 uL ddH2O to each microfuge tube
- Using pipette tip, dip into colonies of interest then dip into labelled microfuge tubes and swirl around to release cells
- Use the the ddH2O+cell mixture as the DNA template for a PCR - instead of adding plain ddH2O to the PCR to bring it up to volume, add the ddH2O+cell mixture
- Confirm your PCR had desired results with a DNA gel, then use the ddH2O+cell mixture from the corresponding microfuge tube the PCR was done on to inoculate a culture or plate.
August 25th, Saturday
Conjugating nah into Shewy:
Caleb, Tina, and a guest Louis ran a gel of the miniprep and colony parallel Phusion PCRs. All of the miniprep PCRs worked, but only a couple colony PCRs worked. We believe this is due to not having added enough cells to the colony PCR tube because some of the colonies were very tiny.
August 26th-31st
August 26th, Sunday
Conjugating nah into Shewy:
Caleb miniprepped two plasmids - one contained the nah operon preceded with a constitutive promoter and the other is to become a backbone containing the origin of transfer required for conjugation and a gene conveying chloramphenicol resistance. The purified plasmids were digested to extract the nah operon and to cut open the backbone - these digests were then run on a gel. Tina gel extracted the backbone then accidentally threw away the rest of the gel that contained the nah operon fragments.
August 27th, Monday
Conjugating nah into Shewy:
Caleb gel extracted the nah operon containing fragments from yesterday - despite having been in the garbage overnight, the gel extraction worked! Caleb and Tina began a 24 hour 16 degrees Celsius ligation using molar rations of nah to backbone of 3:1 and 1:1.
August 28th, Tuesday
Conjugating nah into Shewy:
Caleb and Tina de-salted the 3:1 and 1:1 ligations, then transformed them into a DAP-requiring conjugation strain WM3064 via electroporation. The electroporated cells were plated on DAP + Cm plates.
August 29th, Wednesday
Conjugating nah into Shewy:
Caleb checked the transformation plates after ~14 hours in a 37 degrees Celsius incubator and noticed colonies, but the colonies were barely visible indicating very slow growth of the cells.
August 30th, Thursday
Conjugating nah into Shewy:
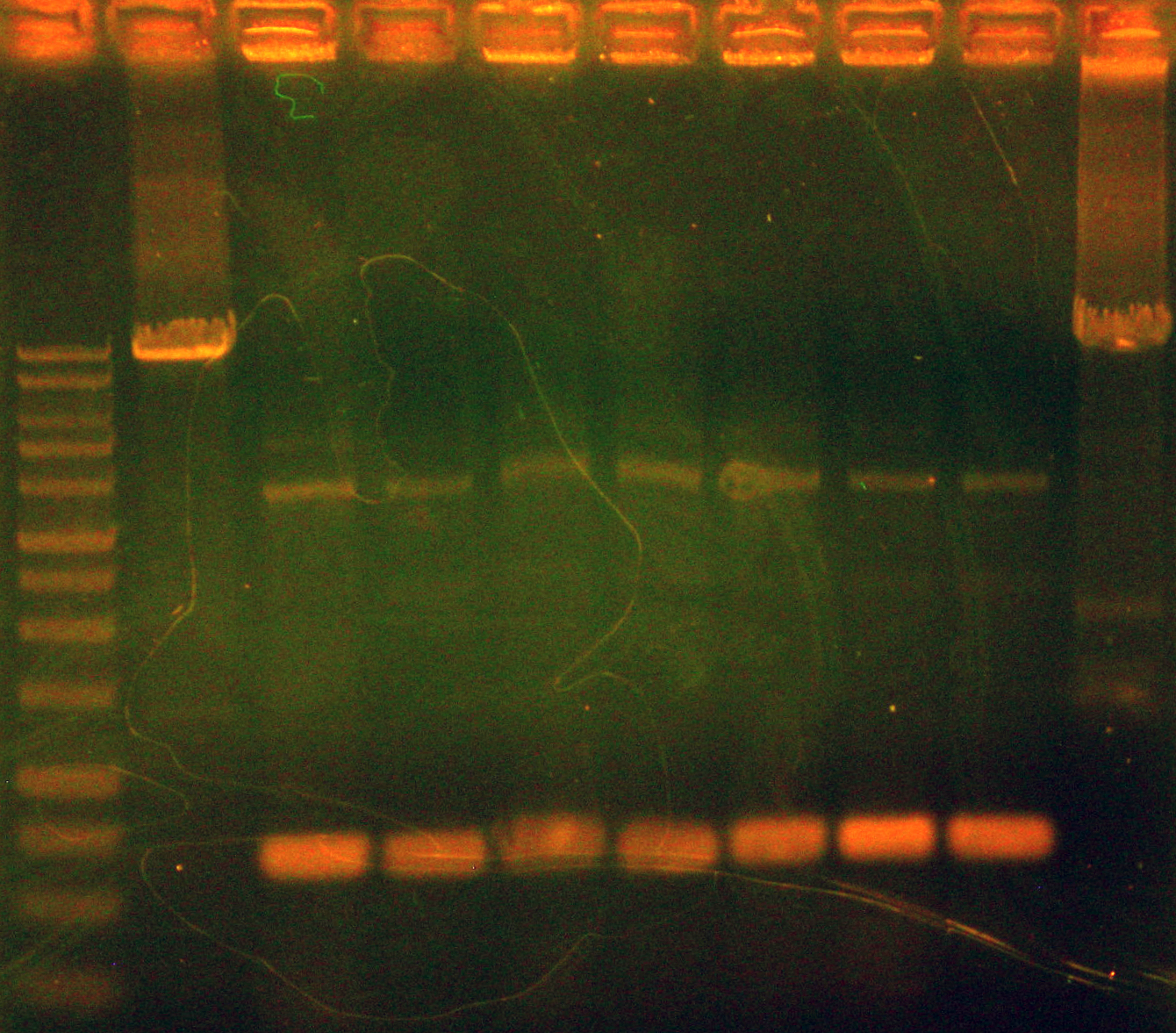
Tina checked the growth of yesterday's colonies and tried doing a colony PCR of each colony and a PCR of a positive control (same product, same primes, but in a different purified plasmid). The primers were designed to sit on the backbone and face into the oriT region and the site where we wanted to insert the nah operon. A PCR amplicon indicating successful ligation of the nah operon into the backbone would be approximately 10-11kb, while a failed ligation would yield a ~700 bp amplicon. Caleb later ran a DNA gel of the colony PCRs and nothing but smears showed up in every lane of the gel except the positive control which didn't even have a smear (Caleb was so disappointed that he didn't post the picture). Since the positive control didn't show up, we concluded the PCR failed. Caleb started overnight cultures of the remaining cells+ddH2O used for the colony PCR.
August 31st, Friday
Conjugating nah into Shewy:
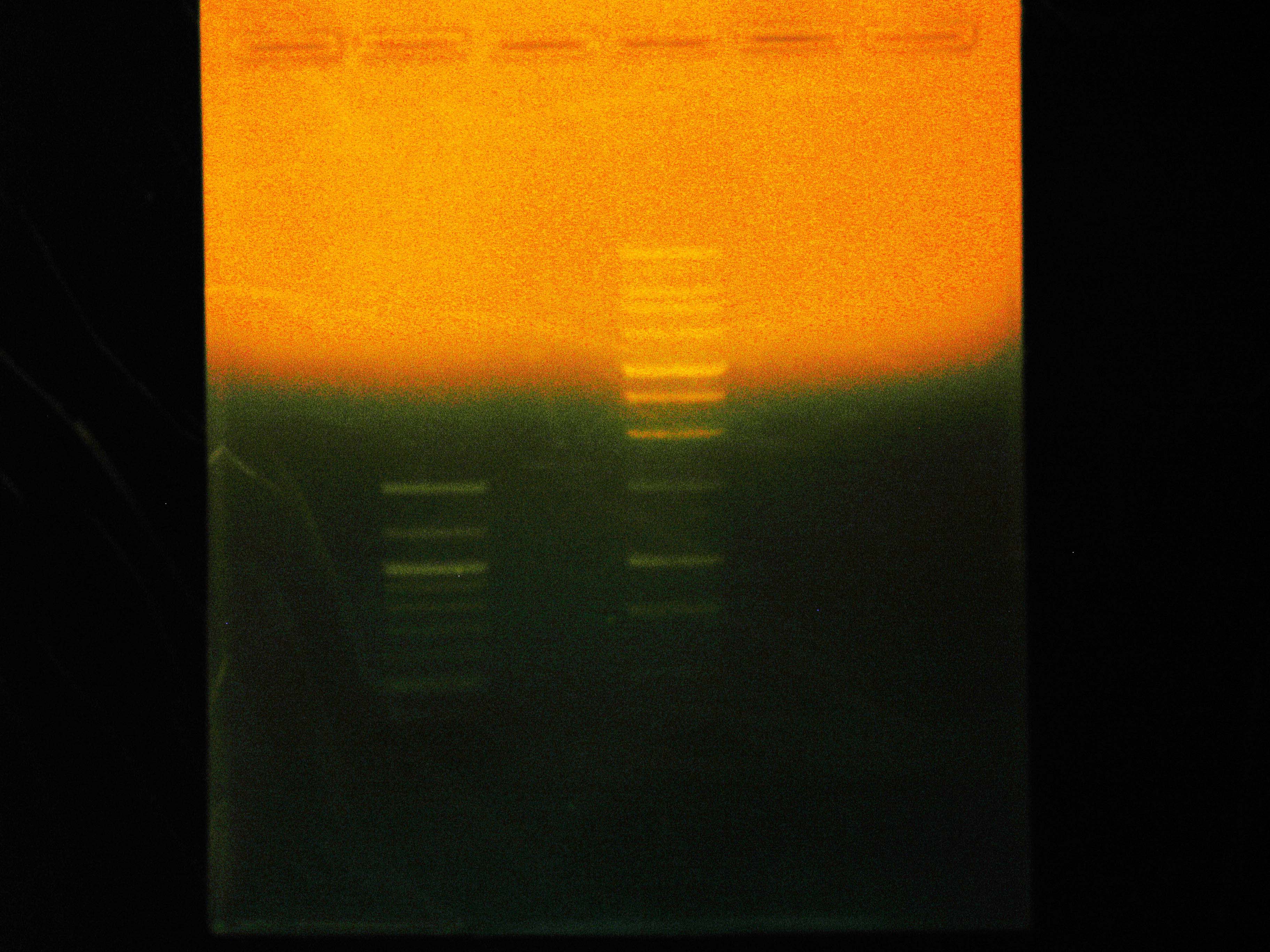
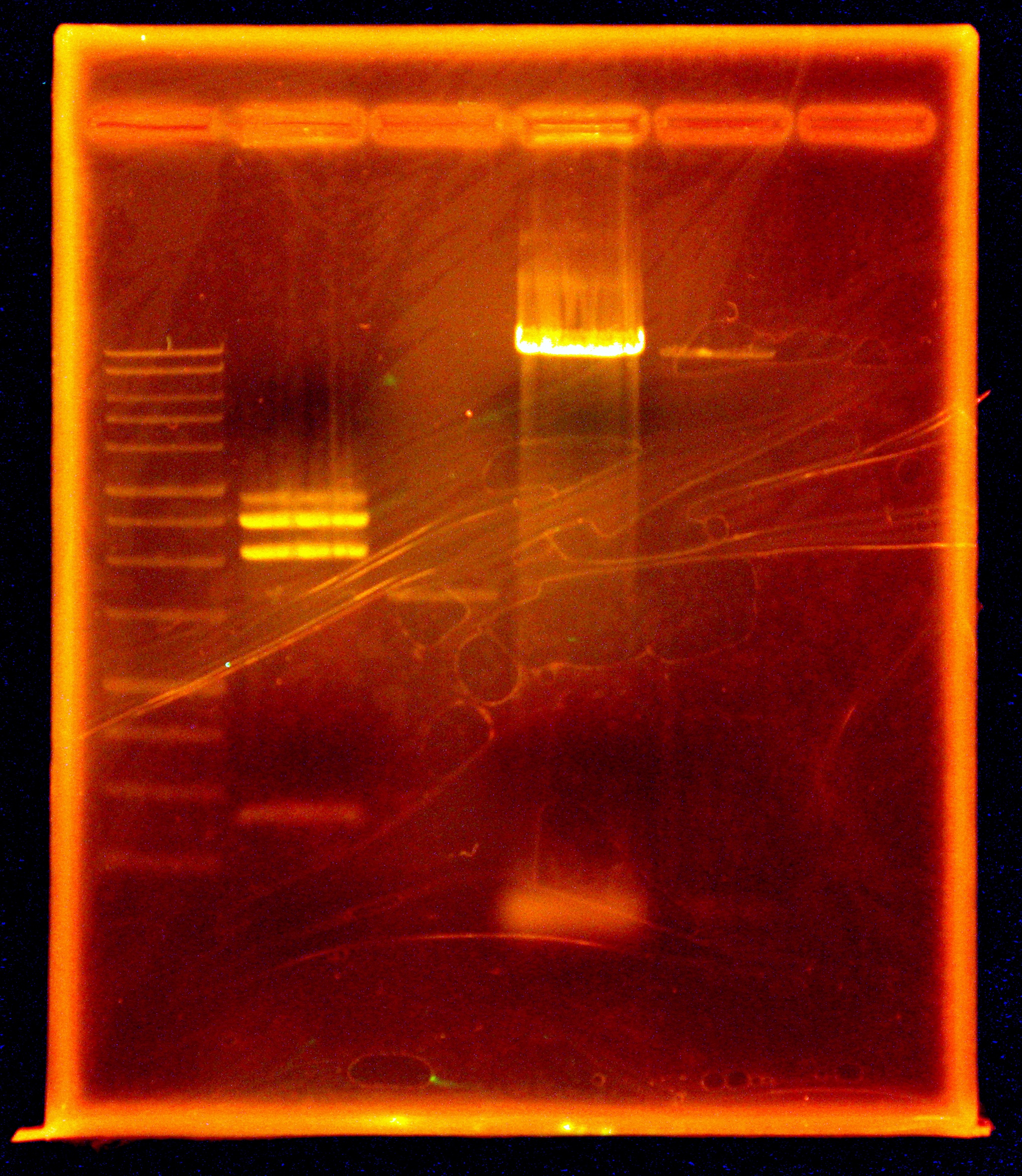
Caleb noticed the overnight cultures didn't grow and concluded the cells must have died due to the cells being stored in an extremely hypotonic solution (ddH2O). He re-did colony PCRs of the colonies, a colony with a positive control, and a positive control of purified plasmid, but modified the colony PCR protocol. Instead of using the cells + ddH2O as template DNA, Caleb dipped the pipette tip into the colony, then dip and swirled it in the PCR rxn tube before dip and swirling into labeled microfuge tubes with 50 uL LB media. The microfuge tubes were stored in a 4 degrees Celsius fridge. A DNA gel of the colony PCRs showed that the ligation of nah into the backbone failed - as seen in the accompanying gel picture, our positive control worked (indicating the PCR conditions were fine), but the test colonies all only had 700 bp amplicons.
September
September 1st-8th
September 1st
Conjugating nah into Shewy:
Due to the mysterious failings of our attempts to ligate the nah operon, Tina tried to start fresh by re-digesting the nah operon-containing plasmid and the oriT-containing backbone.
Synthetic River Media:
Mark and Chie created synthetic river media based on information on the mineral content of the Athabasca River, and started overnight cultures of numerous strains to test how sodium lactate supplemented river water would grow the cells.
September 2nd
Conjugating nah into Shewy:
Tina tried ligating an older digestion of the nah operon into the oriT backbone using a molar ratio or 3:1 and 6:1 for nah:backbone. She then de-salted and transformed the ligations into conjugation strain WM3064. Tina then ran a gel of the digested nah operon-containing plasmid and the oriT-containing backbone and extracted the nah operon and oriT backbone bands.
Synthetic River Media:
Mark set up a 96-well plate to test growth of the engineered strains in sodium lactate at varying concentrations, in synthetic river media.
September 3rd
Conjugating nah into Shewy:
Tina extracted the nah operon digest and the oriT backbone from yesterday's gel. Tina and Dylan had an epiphany - we forgot to dephosphorylate the backbone! No wonder we kept seeing self-ligations! With a renewed spirit, Tina dephosphorylated the digested oriT backbone then started a sixteen hour 16 degrees Celsius ligation of the extracted digests. Tina tried nah to backbone ratios of 1:1, 3:1, and 6:1.
Synthetic River Media:
Mark returned to the lab to discover that the growth assay went wrong. Numerous issues were identified with the original protocol to explain the failed result, and the protocol was changed.
September 4th
Conjugating nah into Shewy:
Tina desalted yesterday's ligations then transformed them into our conjugation strain WM3064. WM3064 is E. coli that is auxotrophic for DAP (diaminopimelic acid epimerase).
September 5th
Conjugating nah into Shewy:
Caleb and Tina checked the transformation plates - some colonies were visible on all three ligation ratio pltaes, but the colonies were so tiny we decided to wait another day before trying colony PCRs or starting liquid cultures.
September 6th
Conjugating nah into Shewy:
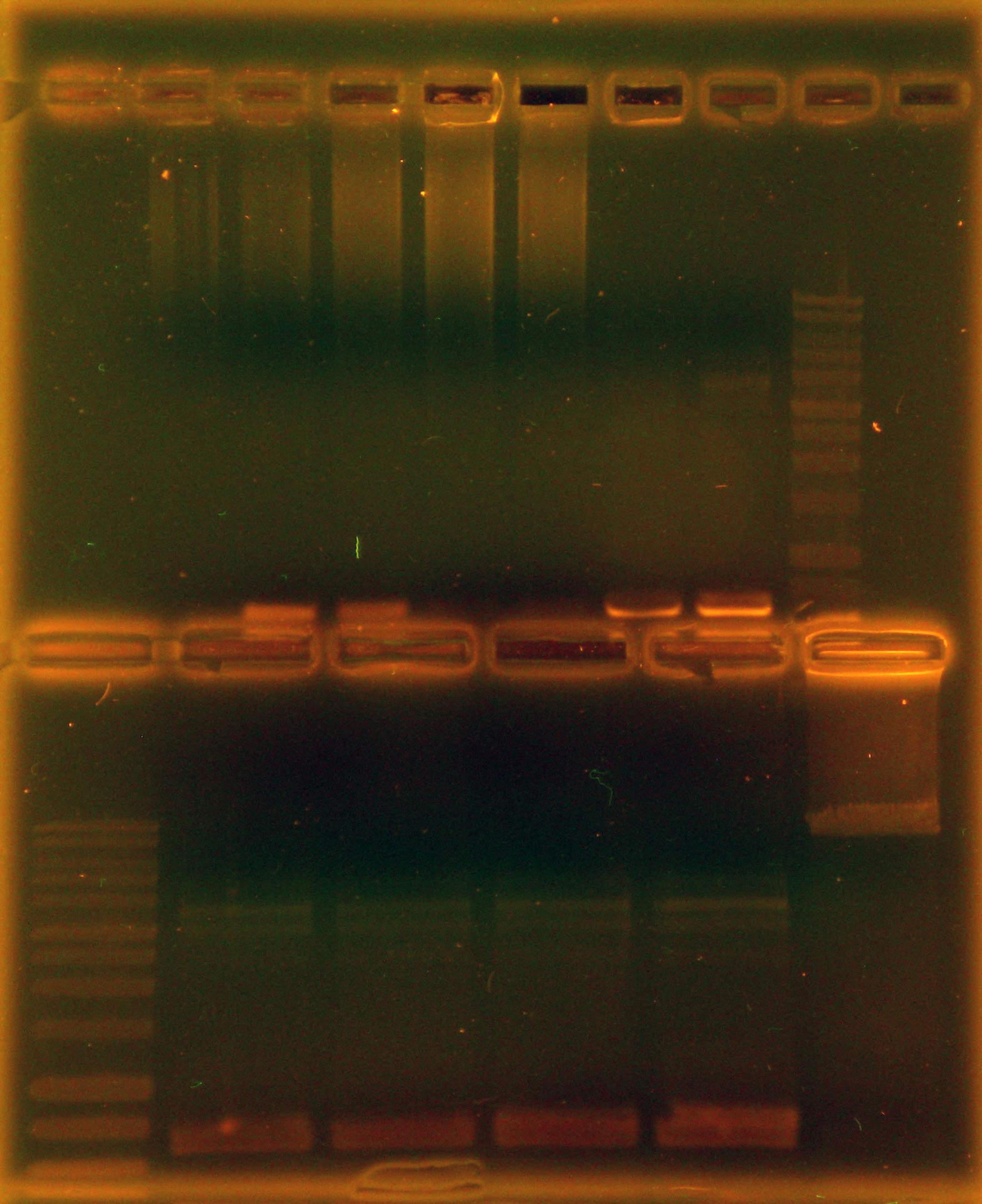
Caleb decided the transformation colonies were large enough to colony PCR and start liquid cultures from. 13 total colony PCRs were performed on colonies from all three ligation mixtures and one positive control was run in parallel to be sure the PCR conditions were correct. Tina ran a gel of the the colony PCRs and the poitive control.
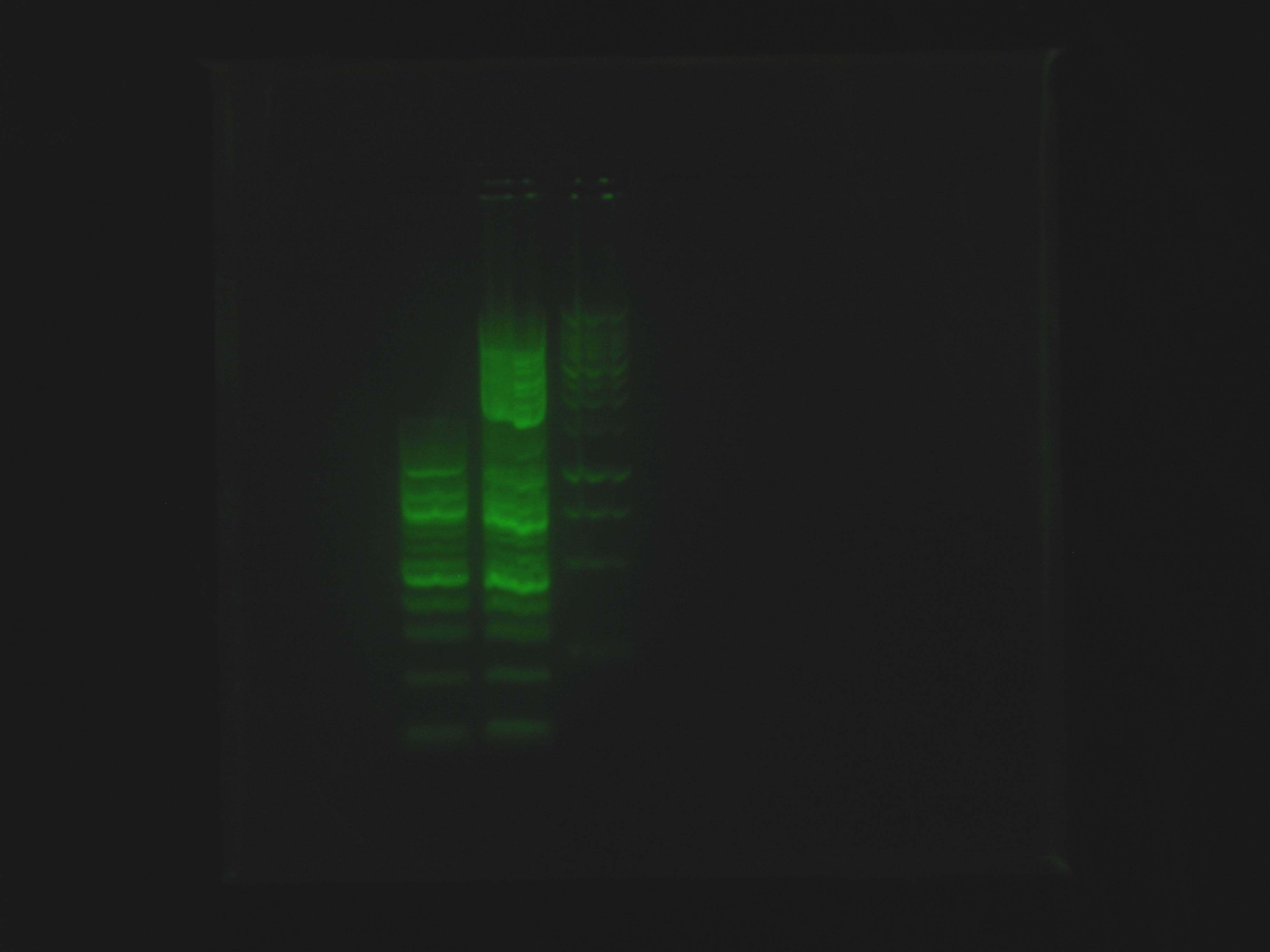
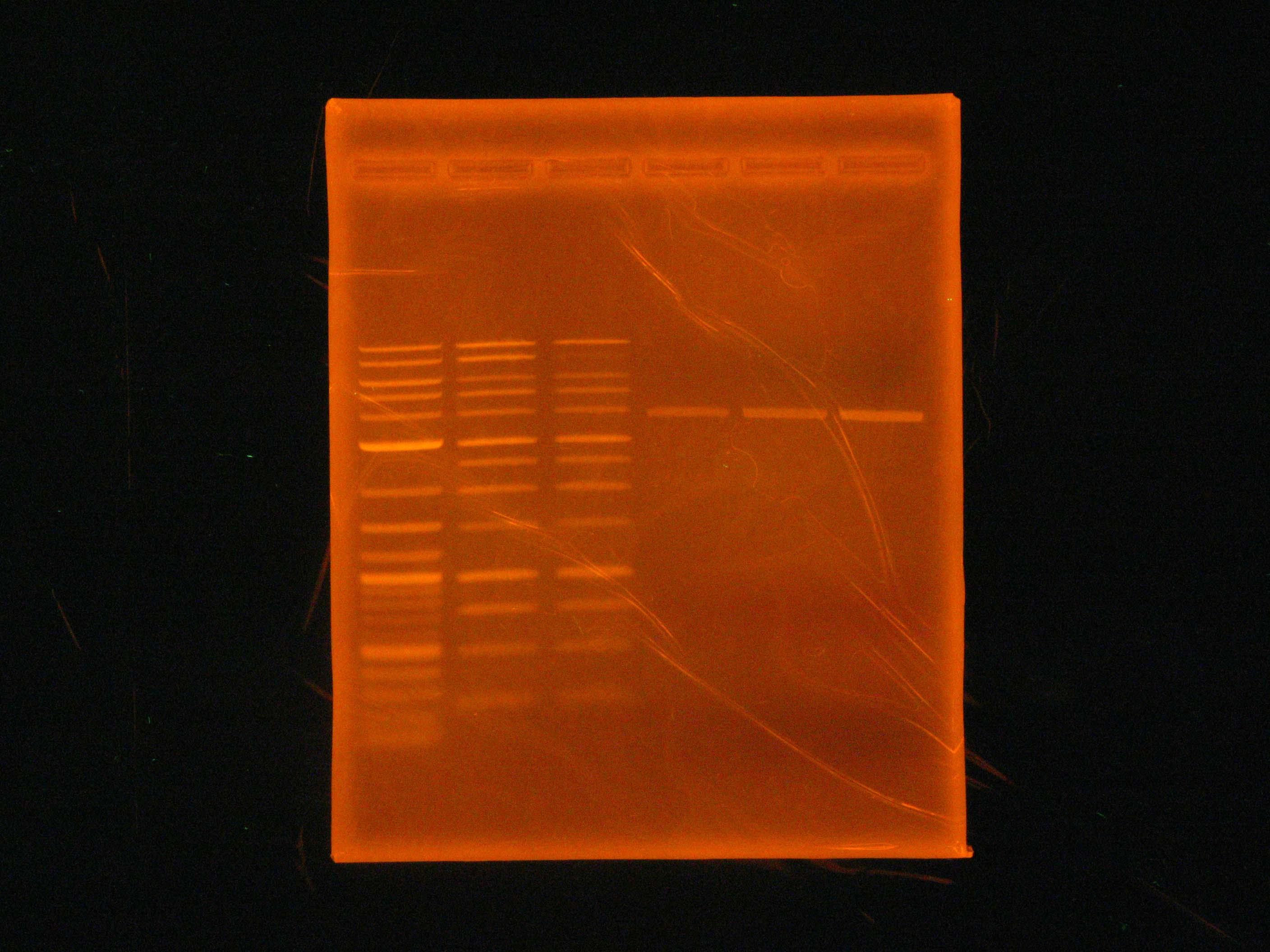
As can be seen in the gel pictures, colonies 3, 4, and 8 definitely had the nah operon and many other colonies had matching middle bands. These matching middle bands likely indicate some sort of mispriming. Colony 5 was clearly the product of an oriT backbone self-ligation. Caleb and Tina started 15 mL liquid cultures of apparently nah containing colonies 3, 4, and 8 and non-nah-containing colony 5 to miniprep from tomorrow.
September 7th
Conjugating nah into Shewy:
Overnight cultures from yesterday looked slightly opaque, but not opaque enough to miniprep from, so Caleb subcultured the cultures hoping the cells would divide fast enough to be ready for miniprepping tomorrow.
Synthetic River Media:
Mark started overnight cultures of all engineered strains to test, again, how growth in synthetic Athabasca river water supplemented with sodium lactate.
September 8th
Conjugating nah into Shewy:
The subcultures of colonies 3, 4, and 8 were not as opaque as Caleb was accustomed to miniprepping from (colony 5 was very opaque, though), but he went ahead and miniprepped anyway because the cultures were already more than 24 hours old. General convention is to miniprep E. coli cultures after ~16 hours.
September 9th-15th
September 9th
Conjugating nah into Shewy:
Caleb checked the miniprep yields today - all but colony 5 had yields below 90 ug/uL. Colony 5 had a yield of over 500 ug/uL. Our hypothesis as to why the E. coli was growing so slowly was that the E. coli was stressed from having to express the many nah operon proteins. Evidence suuporting our hypothesis includes - colony 5 had a much larger colony, grew in liquid much faster, came out as lacking the nah operon, and yielded a lot more DNA from miniprepping than the colonies that supposedly had the nah operon.
September 12th
Conjugating nah into Shewy:
Caleb and Tina started cultures of colony #3 from original transformation plate and cultures of JG700 + SAL and JG700. JG700 is Shewanella oneidensis with a ΔmtrB genotype. SAL refers to a plasmid with our reporter system that responds to the presence of salicylate by upregulating expression of mtrB.
September 13th
Conjugating nah into Shewy:
Caleb started liquid cultures of colonies #3 and #4 from the original transformation plate. Caleb also started a conjugation of JG700 + SAL with colony #3 and JG700 + colony #3. The conjugation was at room temperature on a DAP plate for 16.5 hours.
September 14th
Conjugating nah into Shewy:
Caleb checked the liquid cultures from yesterday and noted there was no growth, as expected, due to the nah operon stressing the cells. Caleb streaked new plates from the conjugation - the LB agar plates had kanamycin and chloramphenicol for the JG700 + SAL and colony #3 conjugation and just chloramphenicol for the JG700 and colony #3 conjugation. The plasmid in the SAL strain confers kanamycin resistance and the plasmid in colony #3 with the nah operon confers chloramphenicol resistance. The strain of E. coli in colony #3 was auxotrophic for DAP, so the E. coli couldn't grow on the newly streaked plates.
September 16th-22nd
September 16th
Conjugating nah into Shewy:
Caleb checked the streaked conjugation plates - there were some colonies, but they were all on top of the initial streak. He and Tina started liquid cultures of all 15 colonies.
September 17th
Conjugating nah into Shewy:
Caleb checked the liquid cultures started yesterday - no growth.
September 18th
Conjugating nah into Shewy:
Caleb checked the liquid cultures started on the 16th - no growth.
September 19th
Conjugating nah into Shewy:
Caleb checked the liquid cultures started on the 16th - no growth.
September 20th
Conjugating nah into Shewy:
Caleb checked the liquid cultures started on the 16th - no growth.
September 21st
Conjugating nah into Shewy:
Caleb checked the liquid cultures started on the 16th - no growth. He concluded that the conjugation failed. Caleb and Tina electroporated the 3:1 (nah:oriT backbone) ligation of dephosphorylated and digested nah with the oriT backbone from September 3rd into both WM3064 and a DH5α strain. They then streaked 16 colonies from the September 4th transformation onto fresh plates.
September 22nd
Conjugating nah into Shewy:
Caleb and Tina noted the appearance of the re-streaked plates - every single streak had numerous colonies. They started liquid cultures and started an overnight colony PCR of all 16 of them. Included with the colony PCRs was a positive control that we knew would have the same amplicon from the nah operon if the colonies were the result of a successful ligation. The electroporated WM3064 plate grew with numerous isolated colonies; the DH5α transformation failed. We hypothesized the DH5α cells were at fault due to their appearance before electroporation - white precipitate was floating in the cell mixture.
September 23rd-30th
September 23rd
Conjugating nah into Shewy:
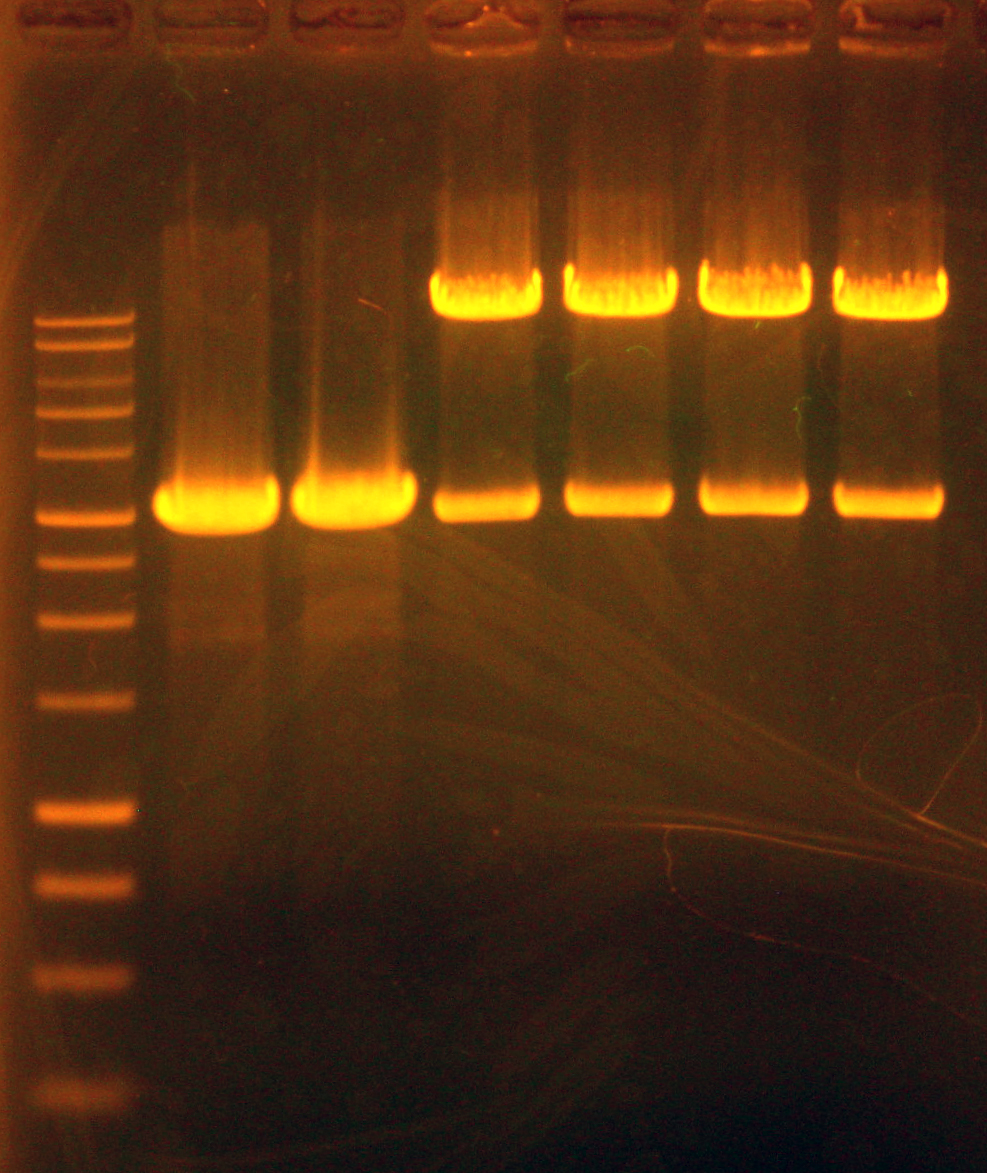
Caleb and Tina ran a gel of the colony PCRs and positive control PCR. "
"