Team:Cambridge/Project/DesignProcess
From 2012.igem.org
| Line 142: | Line 142: | ||
[[File:Lux construct Cam.png|700px|center|thumb|The design and intended function of our ratiometric luciferase construct]]. | [[File:Lux construct Cam.png|700px|center|thumb|The design and intended function of our ratiometric luciferase construct]]. | ||
<html><h3><span class="mw-headline" id="Instrument"></span></h3></html> | <html><h3><span class="mw-headline" id="Instrument"></span></h3></html> | ||
| + | |||
Latest revision as of 03:27, 27 September 2012


Contents |
Design Process
We approached our project through a typical design process, working from our initial project aim and objectives, through market research, investigations, to our implementation and solution. This page contains a more detailed account of our project throughout the summer.
Aim
Every good design process must have a clear aim. Ours was to take a step towards realising the full potential of a group of well characterised parts, the biosensors available within the registry, by developing a prototype kit which could be used effectively and practically in real-world applications. In other words, we aimed to engineer an easy to use product, centred around biobricks, and aimed at the end user. Our team has realised this potential and have developed a kit which has real world uses now, as well as potential applications in the future.
Market Research
Having decided that the primary aim of our project was the design of a practical useful product, the first thing we needed was to find its potential use as well as a possible market. The ideal market would fulfill at least three requirements:
- Face a real problem which could be solved by our product.
- Have the financial capacity to obtain/create our product.
- Be currently in the state where access to similar equipment is limited, and therefore the impact of our solution would be maximised
We aimed to tackle issues concerning the welfare and standard of living of people, in particular those who might lack the resources to tackle them themselves.
To help inform the overall shape of our project, as well as the specifics of our design, we talked to Konrad Siegfried, a scientist who has already had the chance to test a bioreporter construct in the field.
More details about our market research can be found on our Human Practices page
Objectives
As stated in our project aims, and justified by the market reasearch above, we wanted to develop a standard for biosensors that is reliable, affordable, portable, open-source, accessible and relevant. These different targets required further condensation before a formal design process can begin. The formal design goals for our project were, therefore, the development of:
- an output that is reliable, reproducible, and quantitative.
- a robust, standard platform for biosensing inputs.
- a system for the storage and distribution of biosensors for straightforward use in multiple applications.
- a system that is rooted in real world applications that can bring about a positive change in the world
Design considerations
Inputs
It's common sense that without a reliable input system, output systems, however robust, cannot be trusted to give meaningful results. It is therefore important that the response to a given input from a biosensor is well characterised (with response curves) under different conditions. However, to truly revolutionise biosensing we would also require a standard platform which could sense a multitude of analytes. This would allow for better characterisation and all additional information about a sensor's function under different conditions to be acquired with one fewer parameter which would need to be changed. We would also like the response curves to be relatively linear across a wide range such that different toxicity thresholds are detectable using the same system. Ideally, we would also like a system that could be designed "from scratch" in the future such that there would be no need for an existing response mechanism in an organism to be found before implementation as a biosensor could take place. This goal is a very long way off, but we would like to consider platforms with the potential for it. The input must therefore have the following attributes:
- A decoupling between the sensing mechanism and its interface with the output system.
- Highly specific analyte discrimination.
- An output response which is highly proportional to the concentration of analyte present.
- The potential for rational redesign, possibly through the use of software tools.
Processing
All biosensors currently in the registry only use a single output to relay information about the concentration of an analyte. Because there is great potential for differences in culture homogeneity, and because the productivity of cultures is proportional to their cell density, this often leads to poorly reproducible and unreliable results. We therefore desire some processing to be performed (without specifying where in the whole system, yet) which factors in these variables and allows for a much more accurate readout. With accuracy also comes the ability to provide more quantified readouts, assuming the system is capable of the linear response required for such readings. We also want to be able to tune the linear response range based on the end users' desired implementation, in order to further improve usefulness. The processing system must therefore have the following attributes:
- The ability to compensate for culture variability between assays
- The ability to make calculations over a wide range of concentrations with similar, high levels of precision across this range.
Outputs
In order for a project that fits our goals to be commercially successful, it is absolutely vital that a quantitative, numerical, robust, and flexible output exists to relay information to a user that is an accurate representation of the processed input. Outputs fitting this description are commonplace in nearly all other scientific fields where the ability to collect and process data has not only eased interpretation of experiments but also allowed a much faster development of understanding, and ultimately technology, in those fields. In biology however, due to the complexity and inherent variability of the systems under scrutiny, it has been difficult to design such outputs. Additionally, the outputs which exist are either crude, or expensive to produce or interpret. We therefore desire an output with the following characteristics:
- Provides reproducible, quantitative data under many environmental conditions with small error margins.
- Displays data to a user in an intuitive manner that is easy to interpret.
- Be flexible enough to support use in multiple environments
- Does not require specific environmental domains to produce an undistorted post-processing signal.
- Easy to interface with data processing tools
- Capable of supporting multiple readouts from different assays.
- Cheap
Sporage & Distribution
Design considerations must be in place such that our biosensing platform can be distributed and stored easily and cheaply, whether it be for laboratory testing purposes or field applications in places lacking appropriate facilities (for example, areas which cannot keep cultures frozen, or even provide electrical power for a sensor). This relates particularly well to our human practices where we learnt that a major problem to tackle in developing countries is a lack of the conventional biolab infrastructure which most, current technologies rely on.
The design criteria for this aspect of the product are thus as follows:
- Deliverable worldwide
- Can be stored long term without appreciable loss of quality
- Both of the above without requirement for high powered or high tech infrastructure
Investigation and solution
Set out below are the developments the team has made over the summer, in tackling our aim and objectives.
RiboSense
Having decided that we wanted our project to be centred around biosensing, we realised that what we did would need to be compatible with what was already in the registry, as well as any subsequent sensors that people might develop. We therefore went through the registry, and the wikis of the last three years, scouting for sensing parts made by other teams. We found a wide variety of inducible promoters, but very few riboswitch-based sensors. Through our own research, we have found that, whilst there may only be a dozen or so well characterised riboswitches at present, they hold a great deal of potential. We draw this conclusion from the wide availability, and high degree of specificity of the nucleic acid aptamers already on the market. Given that riboswitches are effectively aptamers whose alternative conformations either affect access to regulatory cis sequences, or which add or remove hairpins and other terminator constructs, we believe there is potential for these to be used in the rational design of new riboswitches. We then searched for new riboswitches to characterise and submit and found these two papers: [http://dx.doi.org/10.1126/science.1215063Breaker et al (2012)], and [http://dx.doi.org/10.1016/j.cell.2007.06.051Dann III et al (2007)].
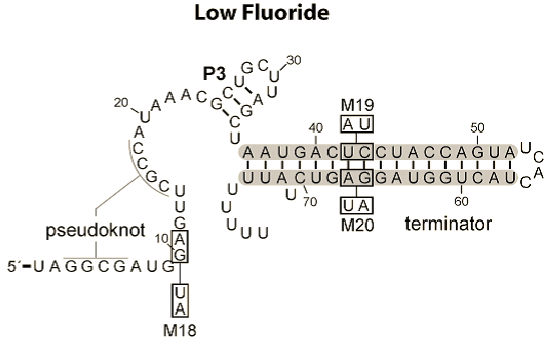
The reason that we chose to work with two riboswitches is because of their differing mechanisms. The fluoride riboswitch turns on its reporter in the presence of fluoride, through the removal of a rho-independent terminator. This is because its in vivo role is to switch on production of a fluoride efflux pump as the [F-] increases above safe thresholds.
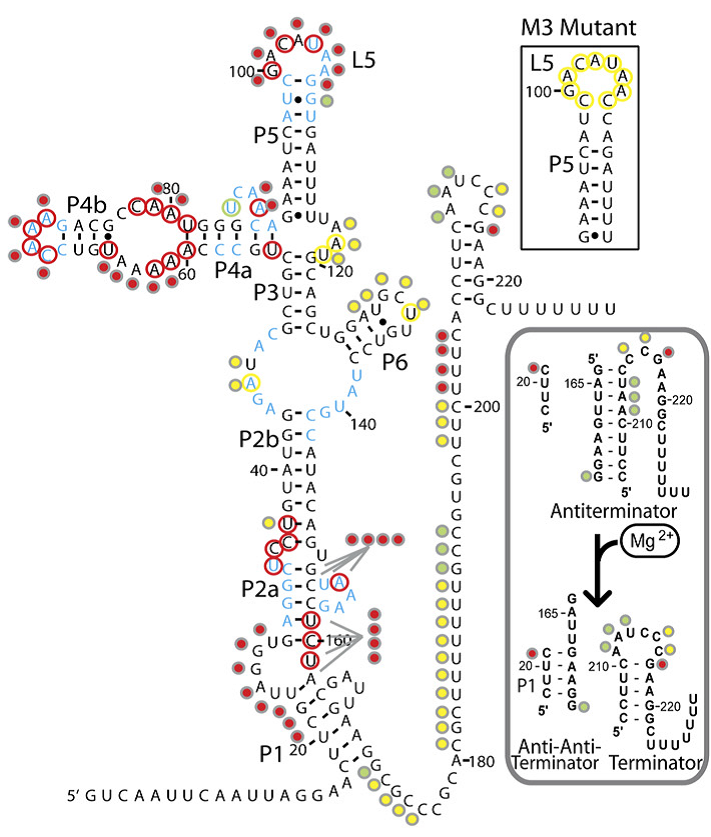
The magnesium riboswitch on the other hand has an influx pump as its effector. Its state in the absence of Mg2+ is to therefore form an antiterminator, this is removed by the anti-antiterminator that forms in the presence of Mg2+.
The logic behind these two choices was that, in order to truly be a universal standard as set out by our design goals above, given that response elements, both promoters and riboswitches, can act to turn a response on or off, our system had to be compatible with both.
Further details on each riboswitch can be found on their respective pages: Fluoride page and Magnesium page
Having identified the riboswitches and designed the constructs to test them we had to obtain the hard DNA to test them. The Magnesium riboswitch is a naturally occurring B.subtilis gene and so was amplified from a sample of genomic DNA previously extracted by PJ Steiner using primers we designed ourselves. The fluoride ribsowitch, however, is a B. cereus gene. Given that this organism is pathogenic we were not permitted to handle it. Therefore, plates of B.subtilis transformed with the gene, as well as a sample of the gene in a shuttle vector, were generously prepared and shipped by members of the Breaker laboratory at Yale University. The Bacillus they sent us lacked the fluoride efflux pump usually turned on by the riboswitch, they found that this greatly increased its sensitivity to fluoride.
We then transformed E.coli and wild type 168 strain B.subtilis with the plasmid they sent us and these three strains were used in parallel for subsequent characterisation. Having obtained the DNA, the first step was to check the construct the fluoride riboswitch was currently in before we eventually would combine it with our own output. For details of these experiments, please see our Results page.
Ratiometrica
We decided to investigate the potential advantages of using dual-channel reporter systems. We anticipate that the inclusion of an internal control signal to which the induced signal can be normalised may significantly reduce variation between results. This would allow more accurate characterisation of the response of a promoter to an inducer, and subsequently would also allow more accurate quantification of an inducer with a readout from the same promoter.
We decided that we would design and assemble two constructs.
The first reporter construct is a dual fluorescence reporter using eCFP and eYFP as the inducible and constitutive reporters respectively. We were advised on this choice by James Brown, who has previously worked on similar constructs. eCFP and eYFP have similar maturation times and stability, and their spectra do not overlap significantly, making them comparable but distinct signals.
Since we were investigating bacillus as a chassis and developing the quick-germination biobrick, we decided to design our construct such that it should function in both e.coli and bacillus.
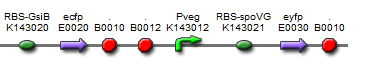
The final design consisted of 4 biobricks ligated into a bacillus shuttle vector, as shown. The promoters pSpac and pVeg, the RBS GsiB and SpoVG are both functional in e.coli and bacillus, giving the construct potential to be tested in both species. pVeg is active constitutively, and pSpac is lacI - induced. For pSpac testing, the shuttle vector includes a constitutively expressed LacI ORF, as bacillus does not naturally express lacI. We intended to assemble in e.coli, characterise and then transform bacillus and further characterise the behaviour of the construct. However, technical difficulties significantly delayed assembly, but we did manage to do some characterisation in E.coli.
For further information, please refer to our Results page.
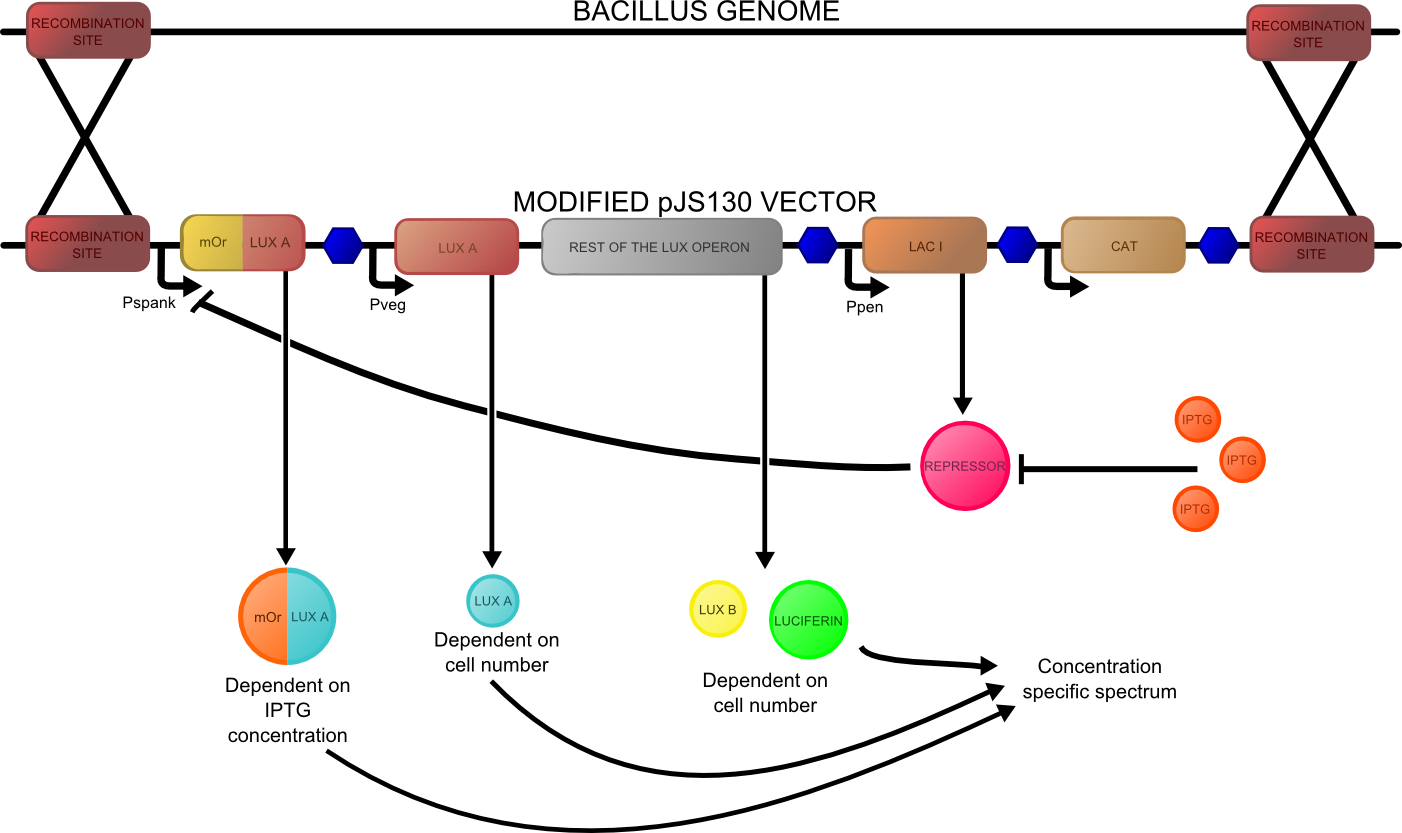
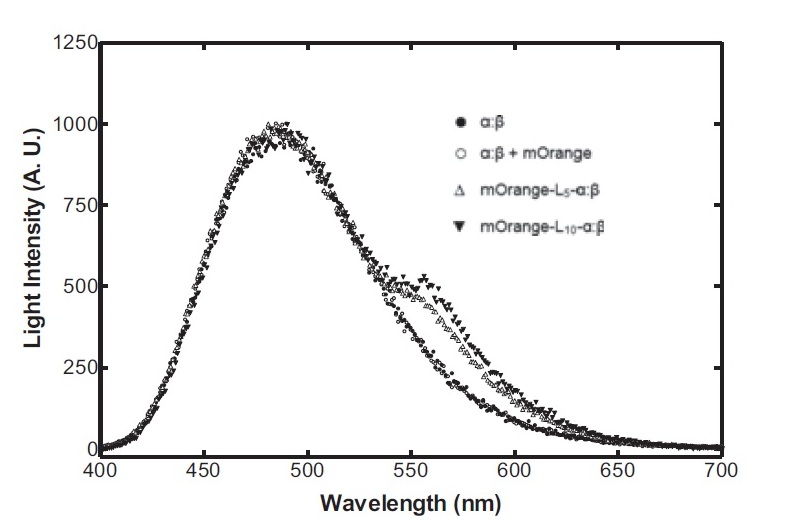
The second reporter construct was designed with our instrumentation in mind. We decided that a luciferase-based output would be the most readily quantifiable with inexpensive components. This left us with the choice of what luciferase(s) to use. Firefly luciferase has been mutagenised into a wide range of colours and would seem the obvious choice. However, it has the flaw of requiring the addition of exogenous luciferin in order to function. Luciferin is expensive, and not stable at room temperature for extended periods of time. Therefore using it would clash with our intentions to take steps towards cheaper and more practical biosensing. Additionally, the brightness would be susceptible to variations in the availability of luciferin to cells, which might introduce errors, which this construct is meant to reduce! Therefore we decided to use bacterial luciferase. The substrate regeneration enzymes for this are known and present in a single operon, and allow long-term light production without addition of substrate. This leaves the question of the second channel, for which we need to develop a colour change variant. Vibrio fischeri Y-1 strain has an accessory protein which shifts the emission spectrum into the yellow, but unfortunately the protein is unstable above 18 C and is therefore impractical. Site-directed mutagenesis of the luxAB complex has not resulted in any drastic colour changes in the literature. However a paper published in 2011 by Dachaun Ke & Shiao-Chun Tu (Photochemistry and Photobiology, 2011, 87: 1346–1353) demonstrated that by fusing mOrange, (a fluorescent protein) to the luxA subunit at the N-terminus via a flexible linker, an additional emission "shoulder" was produced at 560nm, as well as the normal peak at 490 nm. Theoretically, if this luciferase was expressed inducibly and a normal luciferase expressed constitutively, one could take a ratio between 490 and 560 nm as a readout of your input.
We decided to recreate this luciferase fusion by inserting mOrange2 into [http://partsregistry.org/Part:BBa_K325909BBa_K325909] , with the intention of later incorporating that into a construct. In addition, we were given an extremely generous offer of synthesis from DNA 2.0, and we decided that the best use of this offer would be to make a complete reporter construct containing this emission-shifted luciferase. As with the fluorescent construct, this was designed such that it should function in both E.coli and bacillus. As before, pVEG and pSpank were used (although a more sensitive derivative of pSpank, hyperspank was used). Because it would only be present as a single copy when integrated into bacillus, the construct was codon-optimised for bacillus, and strong bacillus RBSes were added, in order to maximise light output.
The final construct is [http://partsregistry.org/wiki/index.php?title=Part:BBa_K911004available on the registry], and characterisation data is available on our results page.
Instrumentation(Biologger)
Having come across the OFP-luciferase fusion, the emission spectra of which appeared sufficiently distinct from that of the normal bacterial luciferase (a fairly distinctive blue), it became clear to the team that this would be a great opportunity for developing our own hardware and software which could be used as part of our kit. The difference in emission spectra could be identified by simple photo-resistors (70kΩ/200kΩ) and coloured theatrical filter gels. The emission spectra of the OFP/luciferase fusion is shown below:
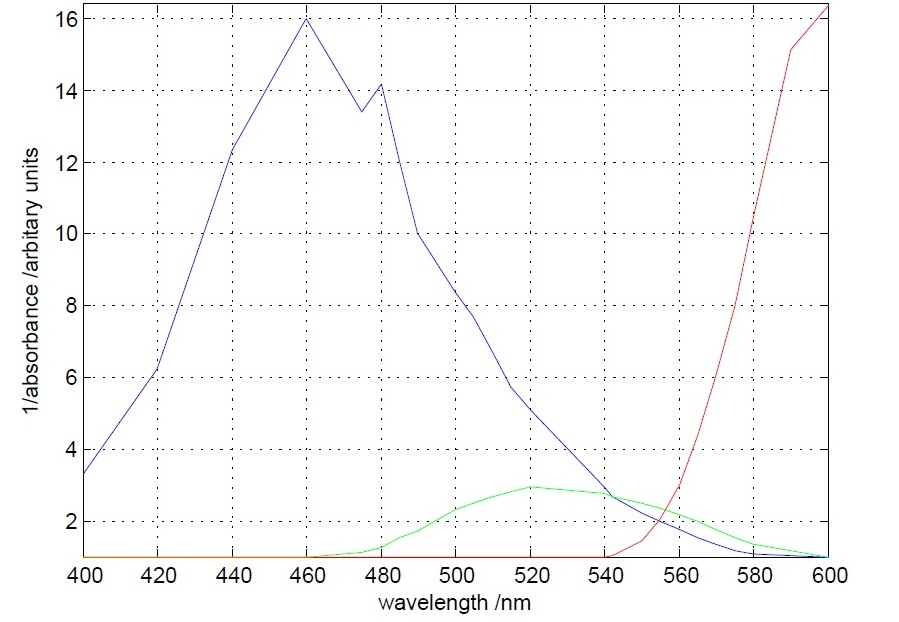
The first experiment to be done was the measurement of the absorbance (actually the inverse of it) of different theatrical filter gels. The target was to differentiate the 560 nm peak to the 490 nm peak. This was accomplished, as it can be seen below, by one blue and one orange filter gel.
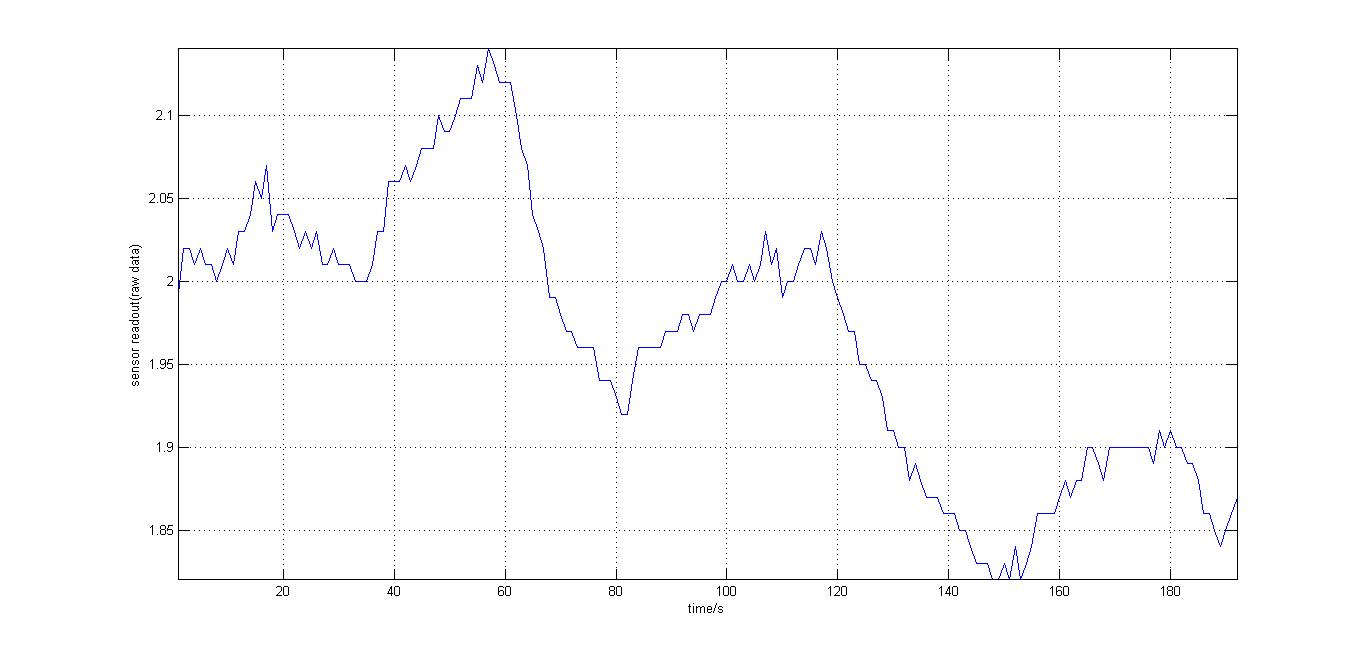
The second experiment to be done was the testing of the sensor with luciferase-producing E.coli with lux genes, taken from the 2010 Cambridge team. The experiment was conducted by moving the tube containing these bacteria towards and away from our sensor (shown on the right). We also tried to use multiple darkness reference readings at different distances from the tube containing the E.coli sto account for a decrease in darkness value over time (probably as the circuit heated up). It should be noted that the orange and blue filters were swapped for this experiment, as compared to our final design. However, this was acceptable at the time as what we wanted to test was whether our sensor could detect bioluminescence. Therefore, light which consisted primarily of blue frequencies caused a rise instead of drop. For all the other experiments the blue and orange filters were placed in the final order. The results were very encouraging, showing that the sensor was certainly reacting to even that minute amount of light.
After we ensured that our idea of developing cheap instrumentation was feasible, we continued with the development of the rest of the instrumentation which included the mechanical, electrical and software parts. What was finally made as well as the reasoning behind it can be found in our Instrumentation (Biologger) page.
The results obtained after the testing of the instrumentation with biological samples as well as videos of our instrumentation in action can be seen at our Results page.
Further improvements on instrumentation design
Due to the constraints of time and budget, we could not realise all of our aims for the Biologger. What we have developed though is a robust prototype which clearly demonstrates the potential our kit has as a cost-effective and reliable sensory ability. We include the considerations we could not realise below, in order to guide anyone who wishes to turn our prototype into a polished end product.
- The cuvette holders we made could be replaced with alternative cuvettes with bio-containment mechanisms. These cuvettes could have a hollow wall, containing an antibiotic or antiseptic. By applying a small amount of force on the sides with the lid on, the second layer would break, thus releasing the antibiotic inside the cuvette. The cuvette would then pose a greatly diminished biocontainment risk and so this would facilitate disposal.
- Microfluidic technology could be used to deliver the testing sample to the cuvettes. This would both reduce spillage, and potentially reduce the sample volumes required.
- For increased biosecurity, and where cost is not an issue, the device can have a sensor dedicated to each cuvette. In that way, parallel readings could be taken in a non-rotary device, reducing the exposure of the cuvettes not in the detector to the environment.
- Hall-effect sensors or barcode readings could be used to more accurately identify which cuvette was in the detector. Furthermore, these barcodes could also be useful for identifying what analyte is being assayed in the sample by each biosensor.
- GPS data could also be acquired by the Android software, thus facilitating data analysis and detection of possible trends related to geographical position.
Sporage and Distribution
Sporulation of 'Bacillus subtilis' cells happens naturally when the organism is stressed. In the lab sporulation can be artificially triggered by growing on a defined medium. The cells are first transformed with all the necessary parts for sensing and output and then sporulated. The dormant spores can be stored by simply dropping a solution of them onto sterile filter paper disks and air drying them. These disks are attached to the inside of the cuvette thereby immobilising the bacteria. A dessicant is also kept in the cuvette to contribute to the longevity. The cuvette can then be kept until the kit is required. When the kit is required, a provided germination medium is added to the cuvette. After a short incubation time the cuvette can be loaded into the arduino device, test solution added, and used for biosensing.
Work Flow
Created with Admarket's flickrSLiDR.
spoVA and Fast Germination
Germination is a time consuming event. The sooner germination and outgrowth is completed, the quicker the biosensing capability of the system can be implemented, which is the primary goal of sporduino.
We came across several works in the literature in modifying aspects of germination. Many referred to the ‘ger’ and ‘spoVA’ operons, the latter or which we investigated further.
In an attempt to increase the rate of germination, we overexpressed the spoVA operon by placing it under the control of the promoter to the ‘B.subtilis’ ‘sspB’ gene (PsspB). PsspB is a germination specific promoter which is more active than the ordinary spoVA promoter.
The construct for this was designed by Peter Setlow’s group, University of Connecticut. It involves the sspB promoter upstream of the first 354bp of the spoVAA member of the spoVA operon. Since ‘B.subtilis’ exhibits accurate and efficient homologous recombination, a single crossover event occurs between the spoVAA 354bp fragment on the construct and the endogenous spoVAA gene in the ‘B.subtilis’ genome, the sspB promoter is inserted upstream of the spoVA operon on the chromosome.
Both the sspB promoter and spoVAA fragment were amplified from ‘B.subtilis’ genomic extracts.
We added the RFC10 standard prefix and suffix to the PsspB/spoVAA construct to make it into the Fast Germination biobrick: BBa_K911008.
 "
"