Team:Paris-Saclay/Project/Notebook/Week 12
From 2012.igem.org
(Difference between revisions)
YohannPetiot (Talk | contribs) |
YohannPetiot (Talk | contribs) |
||
| Line 24: | Line 24: | ||
<body> | <body> | ||
<div id="tiles"> | <div id="tiles"> | ||
| - | + | <div id="single-tile3" class="red live-tile" data-mode="flip" data-delay="4000"> | |
<div> | <div> | ||
<a href="https://2012.igem.org/Team:Paris-Saclay/Project/Abstract"> | <a href="https://2012.igem.org/Team:Paris-Saclay/Project/Abstract"> | ||
<div class="child-tile"><p class="child-tile">GEMOTE Project</p></div> | <div class="child-tile"><p class="child-tile">GEMOTE Project</p></div> | ||
| - | <img src=" | + | <img src="https://static.igem.org/mediawiki/2012/7/7c/Project-1.png" alt="" /> |
</a> | </a> | ||
</div> | </div> | ||
<div> | <div> | ||
<a href="https://2012.igem.org/Team:Paris-Saclay/Project/Abstract"> | <a href="https://2012.igem.org/Team:Paris-Saclay/Project/Abstract"> | ||
| - | <img src=" | + | <img src="https://static.igem.org/mediawiki/2012/d/d1/Project2.jpg" alt="" /> |
<div class="child-tile"><p class="child-tile">GEMOTE Project</p></div> | <div class="child-tile"><p class="child-tile">GEMOTE Project</p></div> | ||
</a> | </a> | ||
Revision as of 22:24, 26 September 2012
Week 12
20th August
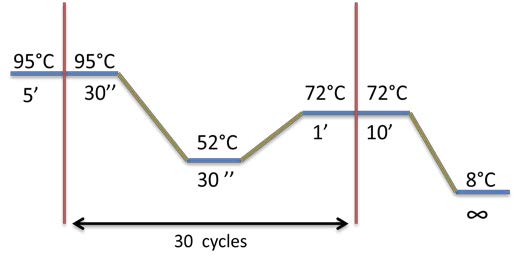
- The 63 colonies obtained after the Gibson for the B construction have been streaked in order to isolate clones. The Petri dishes composed of LB + Ampicilline are placed at 42°C. The #11 colony is the one that turned red.
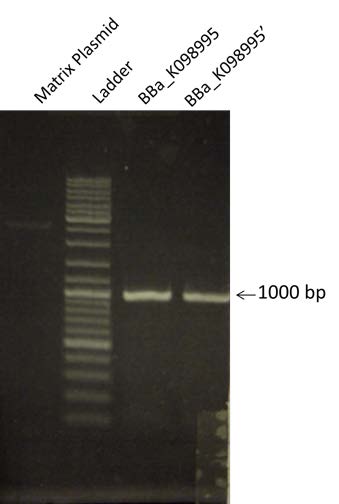
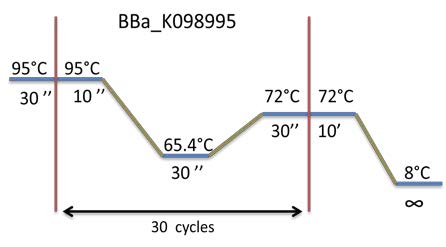
| To prepare the Gibson A construction, BBa_K098995 is amplified by PCR. To verify the Gibson, a 1% Agarose gel Electrophoresis is made. We are expecting a band at a size of 975 bp. The matrix plasmid is a control. |
21th August
- Digestion by DPNI of BBa_K098995 in order to degrade the matrix plasmid.
- Miniprep of the BBa_K098995
22th August
23rd August
| Individual PCR of the 15 candidates + #11 after the PCR on colonies. This time, all the fragments of the Gibson B construction are amplified. Visualization on 0.8% Agarose gel Electrophoresis | 
|
- Streak of the #11 to test if it can grow at 25°C, since we saw that:
- At 37°C, it forms small red colonies
- At 42°C, it forms slowly small red colonies
- At 30°C, it forms normal white colonies
24th August
- Miniprep of the 15 candiates + #11.
- Digestion by AseI of the 15 candidates + #11 in order to verify that each colony has the plasmid with the Gibson B construction.
- PCR to amplify the 880bp end of BBa_K098995 enclosed in the 15 candidate colonies + #11.
 "
"




Follow us !