Team:SJTU-BioX-Shanghai/Project/project1.3
From 2012.igem.org
(Difference between revisions)
AleAlejandro (Talk | contribs) (→Transcription Signal) |
AleAlejandro (Talk | contribs) (→Membrane Rudder) |
||
| Line 54: | Line 54: | ||
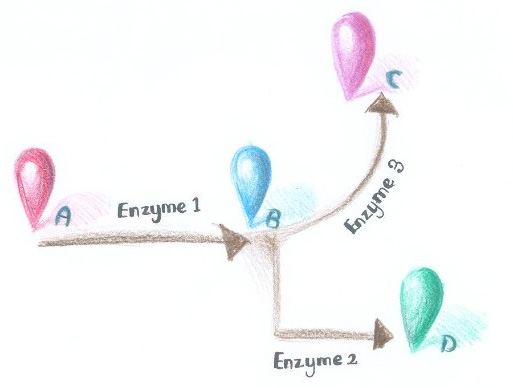
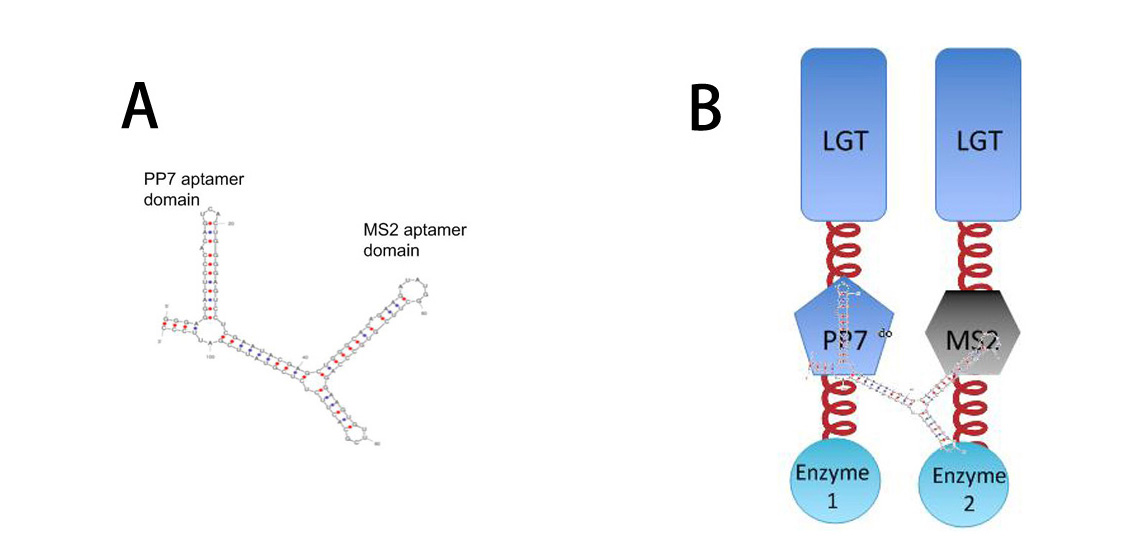
More strikingly, by now, if we want to assemble certain proteins through external signal, we need to find corresponding signal-induced dimer or oligomer. But if we place RNA D0 (with PP7 and MS2 aptamer domains) under control of various promoters regulated by different signals, approaches to induce dimerization would be expanded sharply. Thus, ''Membrane Rudder'' could sense much more signals. | More strikingly, by now, if we want to assemble certain proteins through external signal, we need to find corresponding signal-induced dimer or oligomer. But if we place RNA D0 (with PP7 and MS2 aptamer domains) under control of various promoters regulated by different signals, approaches to induce dimerization would be expanded sharply. Thus, ''Membrane Rudder'' could sense much more signals. | ||
| - | ==Chemical Signal== | + | ==Chemical & Peptide Signal== |
| - | There are many ligand induced dimer in organisms. | + | There are many ligand-induced dimer in organisms. ErbB family proteins are typically ligand-induced dimers, and estrogen receptor only forms dimer when estrogen is present. |
Recruiting those signal induced dimers could easily broaden the application field of our system. What we offer is just a universal tool which can be modified for different use. | Recruiting those signal induced dimers could easily broaden the application field of our system. What we offer is just a universal tool which can be modified for different use. | ||
==Summary== | ==Summary== | ||
| + | |||
| + | |||
| + | ==Reference== | ||
<!----------------------------------------------------end---------------------------------> | <!----------------------------------------------------end---------------------------------> | ||
{{Template:12SJTU_footer}} | {{Template:12SJTU_footer}} | ||
Revision as of 21:45, 26 September 2012
 "
"